ePeak: from replicated chromatin profiling data to epigenomic dynamics
- PMID: 35664802
- PMCID: PMC9154330
- DOI: 10.1093/nargab/lqac041
ePeak: from replicated chromatin profiling data to epigenomic dynamics
Abstract
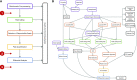
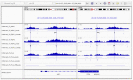
We present ePeak, a Snakemake-based pipeline for the identification and quantification of reproducible peaks from raw ChIP-seq, CUT&RUN and CUT&Tag epigenomic profiling techniques. It also includes a statistical module to perform tailored differential marking and binding analysis with state of the art methods. ePeak streamlines critical steps like the quality assessment of the immunoprecipitation, spike-in calibration and the selection of reproducible peaks between replicates for both narrow and broad peaks. It generates complete reports for data quality control assessment and optimal interpretation of the results. We advocate for a differential analysis that accounts for the biological dynamics of each chromatin factor. Thus, ePeak provides linear and nonlinear methods for normalisation as well as conservative and stringent models for variance estimation and significance testing of the observed marking/binding differences. Using a published ChIP-seq dataset, we show that distinct populations of differentially marked/bound peaks can be identified. We study their dynamics in terms of read coverage and summit position, as well as the expression of the neighbouring genes. We propose that ePeak can be used to measure the richness of the epigenomic landscape underlying a biological process by identifying diverse regulatory regimes.
© The Author(s) 2022. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures

References
-
- Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. - PubMed
-
- Johnson D.S., Mortazavi A., Myers R.M., Wold B.. Genome-wide mapping of in vivo protein-DNA interactions. Science (New York, N.Y.). 2007; 316:1497–1502. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

