Fluorescence spectrophotometry for COVID-19 determination in clinical swab samples
- PMID: 35664893
- PMCID: PMC9150911
- DOI: 10.1016/j.arabjc.2022.104020
Fluorescence spectrophotometry for COVID-19 determination in clinical swab samples
Abstract
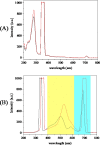
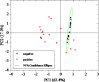
Considering the limitations of the assays currently available for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its emerging variants, a simple and rapid method using fluorescence spectrophotometry was developed to detect coronavirus disease 2019 (COVID-19). Forty clinical swab samples were collected from the nasopharyngeal and oropharyngeal cavities of COVID-19-positive and -negative. Each sample was divided into two parts. The first part of the samples was analyzed using reverse transcription-polymerase chain reaction (RT-qPCR) as the control method to identify COVID-19-positive and -negative samples. The second part of the samples was analyzed using fluorescence spectrophotometry. Fluorescence measurements were performed at excitation and emission wavelengths ranging from 200 to 800 nm. Twenty COVID-19-positive samples and twenty COVID-19-negative samples were detected based on RT-qPCR results. The fluorescence spectrum data indicated that the COVID-19-positive and -negative samples had significantly different characteristics. All positive samples could be distinguished from negative samples by fluorescence spectrophotometry. Principal component analysis showed that COVID-19-positive samples were clustered separately from COVID-19-negative samples. The specificity and accuracy of this experiment reached 100%. Limit of detection (LOD) obtained 42.20 copies/ml (Ct value of 33.65 cycles) for E gene and 63.60 copies/ml (Ct value of 31.36 cycles) for ORF1ab gene. This identification process only required 4 min. Thus, this technique offers an efficient and accurate method to identify an individual with active SARS-CoV-2 infection and can be easily adapted for the early investigation of COVID-19, in general.
Keywords: COVID-19; Emission; Excitation; Fast analysis; Fluorescence spectroscopy; SARS-CoV-2.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
[Evaluation of the Rapid Antigen Detection Kit with the Polymerase Chain Reaction for Detection of SARS-CoV-2 in Respiratory Samples].Mikrobiyol Bul. 2022 Apr;56(2):263-273. doi: 10.5578/mb.20229806. Mikrobiyol Bul. 2022. PMID: 35477229 Turkish.
-
Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test.PLoS One. 2021 Feb 23;16(2):e0243183. doi: 10.1371/journal.pone.0243183. eCollection 2021. PLoS One. 2021. PMID: 33621263 Free PMC article.
-
Diagnostic Performance of Self-Collected Saliva Versus Nasopharyngeal Swab for the Molecular Detection of SARS-CoV-2 in the Clinical Setting.Microbiol Spectr. 2021 Dec 22;9(3):e0046821. doi: 10.1128/Spectrum.00468-21. Epub 2021 Nov 3. Microbiol Spectr. 2021. PMID: 34730436 Free PMC article.
-
Different Respiratory Samples for COVID-19 Detection by Standard and Direct Quantitative RT-PCR: A Literature Review.Iran J Pharm Res. 2021 Summer;20(3):285-299. doi: 10.22037/ijpr.2021.115458.15383. Iran J Pharm Res. 2021. PMID: 34903989 Free PMC article. Review.
-
Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis.J Med Virol. 2021 Feb;93(2):719-725. doi: 10.1002/jmv.26349. Epub 2020 Aug 2. J Med Virol. 2021. PMID: 32706393 Free PMC article.
Cited by
-
Label-free detection of vitamin B by two-step enhanced Raman technique using dynamic borohydride-reduced silver nanoparticles.Mikrochim Acta. 2023 Nov 23;190(12):480. doi: 10.1007/s00604-023-06055-9. Mikrochim Acta. 2023. PMID: 37996711
References
-
- Babady N.E., McMillen T., Jani K., Viale A., Robilotti E.V., Aslam A., Diver M., Sokoli D., Mason G., Shah M.K., Korenstein D., Kamboj M. Performance of severe acute respiratory syndrome coronavirus 2 real-time RT-PCR tests on oral rinses and saliva samples. J. Mol. Diagn. 2021;23:3–9. doi: 10.1016/j.jmoldx.2020.10.018. - DOI - PMC - PubMed
-
- Barauna V.G., Singh M.N., Barbosa L.L., Marcarini W.D., Vassallo P.F., Mill J.G., Ribeiro-Rodrigues R., Warnke P.H., Martin F.L. Ultra-rapid on-site detection of SARS-CoV-2 infection using simple ATR-FTIR spectroscopy and analysis algorithm: high sensitivity and specificity. Anal Chem. 2020;93:2950–2958. doi: 10.1101/2020.11.02.20223560. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
