Single-nucleotide polymorphisms in 3'-untranslated region inducible costimulator gene and the important roles of miRNA in alopecia areata
- PMID: 35664973
- PMCID: PMC9060044
- DOI: 10.1002/ski2.34
Single-nucleotide polymorphisms in 3'-untranslated region inducible costimulator gene and the important roles of miRNA in alopecia areata
Abstract
Background: Alopecia areata (AA) spares the stem cell compartment and attacks only the base of the hair follicle, which is surrounded by infiltrating lymphocytes. AA is associated with polymorphisms in immune-related genes and with decreased function of CD4+CD25+ T regulatory (Treg) cells. Treg function is modulated by the costimulatory molecules, like inducible costimulator (ICOS) that are crucial in orienting T cell differentiation and function so that they strongly impact on the immunologic decision between tolerance or autoimmunity development.
Objective: The aim of our study was to investigate the possible association of AA with single-nucleotide polymorphisms (SNP) present in the ICOS 3'-untranslated region (3'UTR) region and to elucidate how SNPs modulate ICOS gene expression by affecting miRNA binding sites.
Methods: This is a case-control study performed in 184 patients with AA and 200 controls. ICOS gene and miRNA expression were analyzed by real-time polymerase chain reaction.
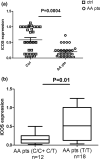
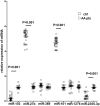
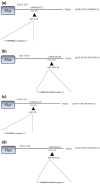
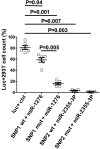
Results: The genotype carrying the rs4404254(C) [p = 0.012, OR (95% CI): 0.5 (0.3-0.8)] and rs4675379(C) [p = 0.015, OR (95% CI): 0.3 (0.1-0.8)] 3' UTR alleles was more frequently observed in AA patients than in controls and correlated with a reduced ICOS expression. miR-1276 significantly suppressed ICOS expression by binding to the 3'UTR of ICOS mRNA. Also, we observed that, miR-101 and miR-27b are upregulated, while miR-103 and miR-2355-3p are downregulated in peripheral blood mononuclear cells of AA patients compared to controls.
Conclusion: Our data show that rs4404254 and rs4675379 SNPs of ICOS gene are associated with AA and also reveal that the presence of rs4404254 polymorphism correlates with ICOS post-transcriptional repression by microRNA binding.
© 2021 The Authors. Skin Health and Disease published by John Wiley & Sons Ltd on behalf of British Association of Dermatologists.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures
References
-
- Safavi KH, Muller SA, Suman VJ, Moshell AN, Melton LJ. Incidence of alopecia areata in Olmsted County, Minnesota, 1975 through 1989. Mayo Clin Proc. 1995;70:628–33. - PubMed
-
- Petukhova L, Cabral RM, Mackay‐Wiggan J, Clynes R, Christiano AM. The genetics of alopecia areata: what's new and how will it help our patients? Dermatol Ther. 2011;24(3):326–36. - PubMed
-
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–87. - PubMed
-
- Sundberg JP, Cordy WR, King LE Jr. Alopecia areata in aging C3H/HeJ mice. J Invest Dermatol. 1994;102:847–56. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
