This is a preprint.
Anti-chemokine antibodies after SARS-CoV-2 infection correlate with favorable disease course
- PMID: 35664993
- PMCID: PMC9164443
- DOI: 10.1101/2022.05.23.493121
Anti-chemokine antibodies after SARS-CoV-2 infection correlate with favorable disease course
Update in
-
Autoantibodies against chemokines post-SARS-CoV-2 infection correlate with disease course.Nat Immunol. 2023 Apr;24(4):604-611. doi: 10.1038/s41590-023-01445-w. Epub 2023 Mar 6. Nat Immunol. 2023. PMID: 36879067 Free PMC article.
Abstract
Infection by SARS-CoV-2 leads to diverse symptoms, which can persist for months. While antiviral antibodies are protective, those targeting interferons and other immune factors are associated with adverse COVID-19 outcomes. Instead, we discovered that antibodies against specific chemokines are omnipresent after COVID-19, associated with favorable disease, and predictive of lack of long COVID symptoms at one year post infection. Anti-chemokine antibodies are present also in HIV-1 infection and autoimmune disorders, but they target different chemokines than those in COVID-19. Monoclonal antibodies derived from COVID- 19 convalescents that bind to the chemokine N-loop impair cell migration. Given the role of chemokines in orchestrating immune cell trafficking, naturally arising anti-chemokine antibodies associated with favorable COVID-19 may be beneficial by modulating the inflammatory response and thus bear therapeutic potential.
One-sentence summary: Naturally arising anti-chemokine antibodies associate with favorable COVID-19 and predict lack of long COVID.
Conflict of interest statement
Figures



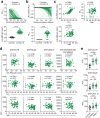


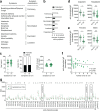
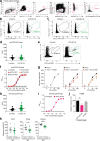

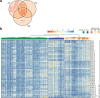



References
-
- Pairo-Castineira E. et al. Genetic mechanisms of critical illness in COVID-19. Nature 591, 92–98 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
