This is a preprint.
The SARS-CoV-2 spike protein binds and modulates estrogen receptors
- PMID: 35665018
- PMCID: PMC9164441
- DOI: 10.1101/2022.05.21.492920
The SARS-CoV-2 spike protein binds and modulates estrogen receptors
Update in
-
The SARS-CoV-2 spike protein binds and modulates estrogen receptors.Sci Adv. 2022 Dec 2;8(48):eadd4150. doi: 10.1126/sciadv.add4150. Epub 2022 Nov 30. Sci Adv. 2022. PMID: 36449624 Free PMC article.
Abstract
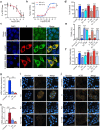
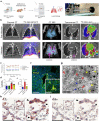
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein binds angiotensin-converting enzyme 2 (ACE2) at the cell surface, which constitutes the primary mechanism driving SARS-CoV-2 infection. Molecular interactions between the transduced S and endogenous proteins likely occur post-infection, but such interactions are not well understood. We used an unbiased primary screen to profile the binding of full-length S against >9,000 human proteins and found significant S-host protein interactions, including one between S and human estrogen receptor alpha (ERα). After confirming this interaction in a secondary assay, we used bioinformatics, supercomputing, and experimental assays to identify a highly conserved and functional nuclear receptor coregulator (NRC) LXD-like motif on the S2 subunit and an S-ERα binding mode. In cultured cells, S DNA transfection increased ERα cytoplasmic accumulation, and S treatment induced ER-dependent biological effects and ACE2 expression. Noninvasive multimodal PET/CT imaging in SARS-CoV-2-infected hamsters using [ 18 F]fluoroestradiol (FES) localized lung pathology with increased ERα lung levels. Postmortem experiments in lung tissues from SARS-CoV-2-infected hamsters and humans confirmed an increase in cytoplasmic ERα expression and its colocalization with S protein in alveolar macrophages. These findings describe the discovery and characterization of a novel S-ERα interaction, imply a role for S as an NRC, and are poised to advance knowledge of SARS-CoV-2 biology, COVID-19 pathology, and mechanisms of sex differences in the pathology of infectious disease.
Figures


References
-
- Mitrovic M. et al. Rotational thromboelastometry (ROTEM) profiling of COVID-19 patients. Platelets 32, 690–696 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
