Mitophagy in Alzheimer's disease: Molecular defects and therapeutic approaches
- PMID: 35665766
- PMCID: PMC9812780
- DOI: 10.1038/s41380-022-01631-6
Mitophagy in Alzheimer's disease: Molecular defects and therapeutic approaches
Abstract
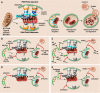
Mitochondrial dysfunctions are central players in Alzheimer's disease (AD). In addition, impairments in mitophagy, the process of selective mitochondrial degradation by autophagy leading to a gradual accumulation of defective mitochondria, have also been reported to occur in AD. We provide an updated overview of the recent discoveries and advancements on mitophagic molecular dysfunctions in AD-derived fluids and cells as well as in AD brains. We discuss studies using AD cellular and animal models that have unraveled the contribution of relevant AD-related proteins (Tau, Aβ, APP-derived fragments and APOE) in mitophagy failure. In accordance with the important role of impaired mitophagy in AD, we report on various therapeutic strategies aiming at stimulating mitophagy in AD and we summarize the benefits of these potential therapeutic strategies in human clinical trials.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

