Reference Genome of the Northwestern Pond Turtle, Actinemys marmorata
- PMID: 35665811
- PMCID: PMC9709993
- DOI: 10.1093/jhered/esac021
Reference Genome of the Northwestern Pond Turtle, Actinemys marmorata
Abstract
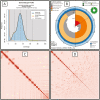
The northwestern pond turtle, Actinemys marmorata, and its recently recognized sister species, the southwestern pond turtle, A. pallida, are the sole aquatic testudines occurring over most of western North America and the only living representatives of the genus Actinemys. Although it historically ranged from Washington state through central California, USA, populations of the northwestern pond turtle have been in decline for decades and the species is afforded state-level protection across its range; it is currently being considered for protection under the US Endangered Species Act. Here, we report a new, chromosome-level assembly of A. marmorata as part of the California Conservation Genomics Project (CCGP). Consistent with the reference genome strategy of the CCGP, we used Pacific Biosciences HiFi long reads and Hi-C chromatin-proximity sequencing technology to produce a de novo assembled genome. The assembly comprises 198 scaffolds spanning 2,319,339,408 base pairs, has a contig N50 of 75 Mb, a scaffold N50 of 146Mb, and BUSCO complete score of 96.7%, making it the most complete testudine assembly of the 24 species from 13 families that are currently available. In combination with the A. pallida reference genome that is currently under construction through the CCGP, the A. marmorata genome will be a powerful tool for documenting landscape genomic diversity, the basis of adaptations to salt tolerance and thermal capacity, and hybridization dynamics between these recently diverged species.
Keywords: CCGP; California Conservation Genomics Project; Emydidae; conservation genetics; testudine.
© The American Genetic Association. 2022.
Figures

References
-
- Agha M, Ennen JR, Bower D, Nowakowski AJ, Sweat S, Todd BD.. 2018. Salinity tolerances and use of saline environments by freshwater turtles: implications of sea level rise. Biol Rev. 93:1634–1648. - PubMed
-
- Bury RB. 2017. Biogeography of western pond turtles in the western Great Basin: dispersal across a Northwest Passage? Western Wildl. 4:72–80.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

