Integrative analysis and prediction of human R-loop binding proteins
- PMID: 35666183
- PMCID: PMC9339281
- DOI: 10.1093/g3journal/jkac142
Integrative analysis and prediction of human R-loop binding proteins
Abstract
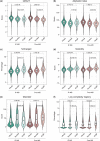
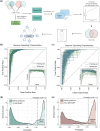
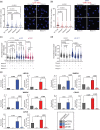
In the past decade, there has been a growing appreciation for R-loop structures as important regulators of the epigenome, telomere maintenance, DNA repair, and replication. Given these numerous functions, dozens, or potentially hundreds, of proteins could serve as direct or indirect regulators of R-loop writing, reading, and erasing. In order to understand common properties shared amongst potential R-loop binding proteins, we mined published proteomic studies and distilled 10 features that were enriched in R-loop binding proteins compared with the rest of the proteome. Applying an easy-ensemble machine learning approach, we used these R-loop binding protein-specific features along with their amino acid composition to create random forest classifiers that predict the likelihood of a protein to bind to R-loops. Known R-loop regulating pathways such as splicing, DNA damage repair and chromatin remodeling are highly enriched in our datasets, and we validate 2 new R-loop binding proteins LIG1 and FXR1 in human cells. Together these datasets provide a reference to pursue analyses of novel R-loop regulatory proteins.
Keywords: R-loop binding proteins; R-loops; random forest.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
