Chromosome-scale assembly with a phased sex-determining region resolves features of early Z and W chromosome differentiation in a wild octoploid strawberry
- PMID: 35666193
- PMCID: PMC9339316
- DOI: 10.1093/g3journal/jkac139
Chromosome-scale assembly with a phased sex-determining region resolves features of early Z and W chromosome differentiation in a wild octoploid strawberry
Abstract
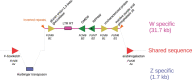
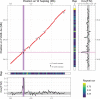
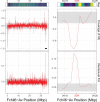
When sex chromosomes stop recombining, they start to accumulate differences. The sex-limited chromosome (Y or W) especially is expected to degenerate via the loss of nucleotide sequence and the accumulation of repetitive sequences. However, how early signs of degeneration can be detected in a new sex chromosome is still unclear. The sex-determining region of the octoploid strawberries is young, small, and dynamic. Using PacBio HiFi reads, we obtained a chromosome-scale assembly of a female (ZW) Fragaria chiloensis plant carrying the youngest and largest of the known sex-determining region on the W in strawberries. We fully characterized the previously incomplete sex-determining region, confirming its gene content, genomic location, and evolutionary history. Resolution of gaps in the previous characterization of the sex-determining region added 10 kb of sequence including a noncanonical long terminal repeat-retrotransposon; whereas the Z sequence revealed a Harbinger transposable element adjoining the sex-determining region insertion site. Limited genetic differentiation of the sex chromosomes coupled with structural variation may indicate an early stage of W degeneration. The sex chromosomes have a similar percentage of repeats but differ in their repeat distribution. Differences in the pattern of repeats (transposable element polymorphism) apparently precede sex chromosome differentiation, thus potentially contributing to recombination cessation as opposed to being a consequence of it.
Keywords: polyploid; sex chromosomes; strawberry; whole-genome assembly.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures



References
-
- Almeida P, Proux-Wera E, Churcher A, Soler L, Dainat J, Pucholt P, Nordlund J, Martin T, Rönnberg-Wästljung A-C, Nystedt B, et al.Genome assembly of the basket willow, Salix viminalis, reveals earliest stages of sex chromosome expansion. BMC Biol. 2020;18(1):78. doi:10.1186/s12915-020-00808-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
