The impact of genetic modifiers on variation in germline mutation rates within and among human populations
- PMID: 35666194
- PMCID: PMC9339295
- DOI: 10.1093/genetics/iyac087
The impact of genetic modifiers on variation in germline mutation rates within and among human populations
Abstract
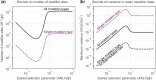
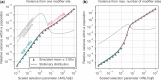
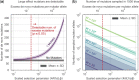
Mutation rates and spectra differ among human populations. Here, we examine whether this variation could be explained by evolution at mutation modifiers. To this end, we consider genetic modifier sites at which mutations, "mutator alleles," increase genome-wide mutation rates and model their evolution under purifying selection due to the additional deleterious mutations that they cause, genetic drift, and demographic processes. We solve the model analytically for a constant population size and characterize how evolution at modifier sites impacts variation in mutation rates within and among populations. We then use simulations to study the effects of modifier sites under a plausible demographic model for Africans and Europeans. When comparing populations that evolve independently, weakly selected modifier sites (2Nes≈1), which evolve slowly, contribute the most to variation in mutation rates. In contrast, when populations recently split from a common ancestral population, strongly selected modifier sites (2Nes≫1), which evolve rapidly, contribute the most to variation between them. Moreover, a modest number of modifier sites (e.g. 10 per mutation type in the standard classification into 96 types) subject to moderate to strong selection (2Nes>1) could account for the variation in mutation rates observed among human populations. If such modifier sites indeed underlie differences among populations, they should also cause variation in mutation rates within populations and their effects should be detectable in pedigree studies.
Keywords: germline mutations; human evolution; modifiers; mutation rate; mutation spectra; mutators; pedigree studies; population genetics.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

