A role for the mitotic proteins Bub3 and BuGZ in transcriptional regulation of catalase-3 expression
- PMID: 35666721
- PMCID: PMC9203020
- DOI: 10.1371/journal.pgen.1010254
A role for the mitotic proteins Bub3 and BuGZ in transcriptional regulation of catalase-3 expression
Abstract
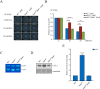
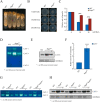
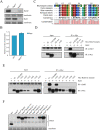
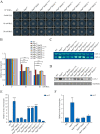
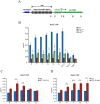
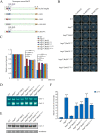
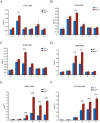
The spindle assembly checkpoint factors Bub3 and BuGZ play critical roles in mitotic process, but little is known about their roles in other cellular processes in eukaryotes. In aerobic organisms, transcriptional regulation of catalase genes in response to developmental or environmental stimuli is necessary for redox homeostasis. Here, we demonstrate that Bub3 and BuGZ negatively regulate cat-3 transcription in the model filamentous fungus Neurospora crassa. The absence of Bub3 caused a significant decrease in BuGZ protein levels. Our data indicate that BuGZ and Bub3 interact directly via the GLEBS domain of BuGZ. Despite loss of the interaction, the amount of BuGZ mutant protein negatively correlated with the cat-3 expression level, indicating that BuGZ amount rather than Bub3-BuGZ interaction determines cat-3 transcription level. Further experiments demonstrated that BuGZ binds directly to the cat-3 gene and responses to cat-3 overexpression induced by oxidative stresses. However, the zinc finger domains of BuGZ have no effects on DNA binding, although mutations of these highly conserved domains lead to loss of cat-3 repression. The deposition of BuGZ along cat-3 chromatin hindered the recruitment of transcription activators GCN4/CPC1 and NC2 complex, thereby preventing the assembly of the transcriptional machinery. Taken together, our results establish a mechanism for how mitotic proteins Bub3 and BuGZ functions in transcriptional regulation in a eukaryotic organism.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
BuGZ facilitates loading of spindle assembly checkpoint proteins to kinetochores in early mitosis.J Biol Chem. 2020 Oct 23;295(43):14666-14677. doi: 10.1074/jbc.RA120.013598. Epub 2020 Aug 20. J Biol Chem. 2020. PMID: 32820050 Free PMC article.
-
A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting.Dev Cell. 2014 Feb 10;28(3):268-81. doi: 10.1016/j.devcel.2013.12.013. Epub 2014 Jan 23. Dev Cell. 2014. PMID: 24462186 Free PMC article.
-
SETD1A function in leukemia is mediated through interaction with mitotic regulators BuGZ/BUB3.EMBO Rep. 2023 Oct 9;24(10):e57108. doi: 10.15252/embr.202357108. Epub 2023 Aug 3. EMBO Rep. 2023. PMID: 37535603 Free PMC article.
-
Research progress of Bub3 gene in malignant tumors.Cell Biol Int. 2022 May;46(5):673-682. doi: 10.1002/cbin.11740. Epub 2022 Feb 24. Cell Biol Int. 2022. PMID: 34882895 Free PMC article. Review.
-
Kinetochore-microtubule interactions "in check" by Bub1, Bub3 and BubR1: The dual task of attaching and signalling.Cell Cycle. 2008 Jun 15;7(12):1763-8. doi: 10.4161/cc.7.12.6180. Epub 2008 Jun 20. Cell Cycle. 2008. PMID: 18594200 Review.
Cited by
-
Nutriepigenomics in Environmental-Associated Oxidative Stress.Antioxidants (Basel). 2023 Mar 21;12(3):771. doi: 10.3390/antiox12030771. Antioxidants (Basel). 2023. PMID: 36979019 Free PMC article. Review.
-
Transcription factor VIB-1 activates catalase-3 expression by promoting PIC assembly in Neurospora crassa.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf174. doi: 10.1093/nar/gkaf174. Nucleic Acids Res. 2025. PMID: 40087884 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

