Regulation of HLA class I expression by non-coding gene variations
- PMID: 35666741
- PMCID: PMC9170083
- DOI: 10.1371/journal.pgen.1010212
Regulation of HLA class I expression by non-coding gene variations
Abstract
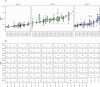
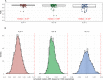
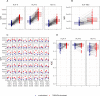
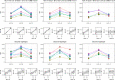
The Human Leukocyte Antigen (HLA) is a critical genetic system for different outcomes after solid organ and hematopoietic cell transplantation. Its polymorphism is usually determined by molecular technologies at the DNA level. A potential role of HLA allelic expression remains under investigation in the context of the allogenic immune response between donors and recipients. In this study, we quantified the allelic expression of all three HLA class I loci (HLA-A, B and C) by RNA sequencing and conducted an analysis of expression quantitative traits loci (eQTL) to investigate whether HLA expression regulation could be associated with non-coding gene variations. HLA-B alleles exhibited the highest expression levels followed by HLA-C and HLA-A alleles. The max fold expression variation was observed for HLA-C alleles. The expression of HLA class I loci of distinct individuals demonstrated a coordinated and paired expression of both alleles of the same locus. Expression of conserved HLA-A~B~C haplotypes differed in distinct PBMC's suggesting an individual regulated expression of both HLA class I alleles and haplotypes. Cytokines TNFα /IFNβ, which induced a very similar upregulation of HLA class I RNA and cell surface expression across alleles did not modify the individually coordinated expression at the three HLA class I loci. By identifying cis eQTLs for the HLA class I genes, we show that the non-coding eQTLs explain 29%, 13%, and 31% of the respective HLA-A, B, C expression variance in unstimulated cells, and 9%, 23%, and 50% of the variance in cytokine-stimulated cells. The eQTLs have significantly higher effect sizes in stimulated cells compared to unstimulated cells for HLA-B and HLA-C genes expression. Our data also suggest that the identified eQTLs are independent from the coding variation which defines HLA alleles and thus may be influential on intra-allele expression variability although they might not represent the causal eQTLs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

