Effects of Inhibitors of SLC9A-Type Sodium-Proton Exchangers on Survival Motor Neuron 2 (SMN2) mRNA Splicing and Expression
- PMID: 35667685
- PMCID: PMC9341265
- DOI: 10.1124/molpharm.122.000529
Effects of Inhibitors of SLC9A-Type Sodium-Proton Exchangers on Survival Motor Neuron 2 (SMN2) mRNA Splicing and Expression
Abstract
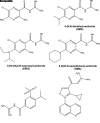
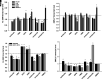
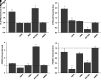
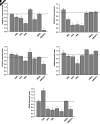
Spinal muscular atrophy (SMA) is an autosomal recessive, pediatric-onset disorder caused by the loss of spinal motor neurons, thereby leading to muscle atrophy. SMA is caused by the loss of or mutations in the survival motor neuron 1 (SMN1) gene. SMN1 is duplicated in humans to give rise to the paralogous survival motor neuron 2 (SMN2) gene. This paralog is nearly identical except for a cytosine to thymine transition within an exonic splicing enhancer element within exon 7. As a result, the majority of SMN2 transcripts lack exon 7 (SMNΔ7), which produces a truncated and unstable SMN protein. Since SMN2 copy number is inversely related to disease severity, it is a well established target for SMA therapeutics development. 5-(N-ethyl-N-isopropyl)amiloride (EIPA), an inhibitor of sodium/proton exchangers (NHEs), has previously been shown to increase exon 7 inclusion and SMN protein levels in SMA cells. In this study, NHE inhibitors were evaluated for their ability to modulate SMN2 expression. EIPA as well as 5-(N,N-hexamethylene)amiloride (HMA) increase exon 7 inclusion in SMN2 splicing reporter lines as well as in SMA fibroblasts. The EIPA-induced exon 7 inclusion occurs via a unique mechanism that does not involve previously identified splicing factors. Transcriptome analysis identified novel targets, including TIA1 and FABP3, for further characterization. EIPA and HMA are more selective at inhibiting the NHE5 isoform, which is expressed in fibroblasts as well as in neuronal cells. These results show that NHE5 inhibition increases SMN2 expression and may be a novel target for therapeutics development. SIGNIFICANCE STATEMENT: This study demonstrates a molecular mechanism by which inhibitors of the sodium-protein exchanger increase the alternative splicing of SMN2 in spinal muscular atrophy cells. NHE5 selective inhibitors increase the inclusion of full-length SMN2 mRNAs by targeting TIA1 and FABP3 expression, which is distinct from other small molecule regulators of SMN2 alternative splicing. This study provides a novel means to increase full-length SMN2 expression and a novel target for therapeutics development.
Copyright © 2022 by The American Society for Pharmacology and Experimental Therapeutics.
Figures



References
-
- Andreassi C, Jarecki J, Zhou J, Coovert DD, Monani UR, Chen X, Whitney M, Pollok B, Zhang M, Androphy E,, et al. (2001) Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet 10:2841–2849. - PubMed
-
- Baranello G, Darras BT, Day JW, Deconinck N, Klein A, Masson R, Mercuri E, Rose K, El-Khairi M, Gerber M,, et al. ; FIREFISH Working Group (2021) Risdiplam in type 1 spinal muscular atrophy. N Engl J Med 384:915–923. - PubMed
-
- Borsi L, Balza E, Gaggero B, Allemanni G, Zardi L (1995) The alternative splicing pattern of the tenascin-C pre-mRNA is controlled by the extracellular pH. J Biol Chem 270:6243–6245. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

