Comprehensive genetic analysis of the human lipidome identifies loci associated with lipid homeostasis with links to coronary artery disease
- PMID: 35668104
- PMCID: PMC9170690
- DOI: 10.1038/s41467-022-30875-7
Comprehensive genetic analysis of the human lipidome identifies loci associated with lipid homeostasis with links to coronary artery disease
Abstract
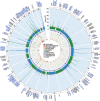
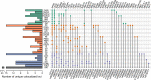
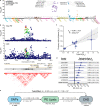
We integrated lipidomics and genomics to unravel the genetic architecture of lipid metabolism and identify genetic variants associated with lipid species putatively in the mechanistic pathway for coronary artery disease (CAD). We quantified 596 lipid species in serum from 4,492 individuals from the Busselton Health Study. The discovery GWAS identified 3,361 independent lipid-loci associations, involving 667 genomic regions (479 previously unreported), with validation in two independent cohorts. A meta-analysis revealed an additional 70 independent genomic regions associated with lipid species. We identified 134 lipid endophenotypes for CAD associated with 186 genomic loci. Associations between independent lipid-loci with coronary atherosclerosis were assessed in ∼456,000 individuals from the UK Biobank. Of the 53 lipid-loci that showed evidence of association (P < 1 × 10-3), 43 loci were associated with at least one lipid endophenotype. These findings illustrate the value of integrative biology to investigate the aetiology of atherosclerosis and CAD, with implications for other complex diseases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG046171/AG/NIA NIH HHS/United States
- R01 AG019771/AG/NIA NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- U01 AG024904/AG/NIA NIH HHS/United States
- U19 AG063744/AG/NIA NIH HHS/United States
- R01 CA129769/CA/NCI NIH HHS/United States
- R01 AG061788/AG/NIA NIH HHS/United States
- RF1 AG059093/AG/NIA NIH HHS/United States
- RF1 AG057452/AG/NIA NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- P30 AG010133/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- R03 AG054936/AG/NIA NIH HHS/United States
- K01 AG049050/AG/NIA NIH HHS/United States
- R01 LM012535/LM/NLM NIH HHS/United States
- U01 AG061359/AG/NIA NIH HHS/United States
- RF1 AG058942/AG/NIA NIH HHS/United States
- RF1 AG051550/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

