The metabolic enzyme hexokinase 2 localizes to the nucleus in AML and normal haematopoietic stem and progenitor cells to maintain stemness
- PMID: 35668135
- PMCID: PMC9203277
- DOI: 10.1038/s41556-022-00925-9
The metabolic enzyme hexokinase 2 localizes to the nucleus in AML and normal haematopoietic stem and progenitor cells to maintain stemness
Abstract
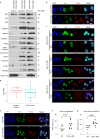
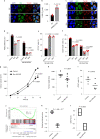
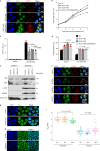
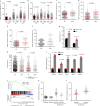
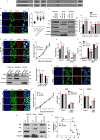
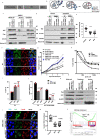
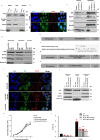
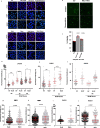
Mitochondrial metabolites regulate leukaemic and normal stem cells by affecting epigenetic marks. How mitochondrial enzymes localize to the nucleus to control stem cell function is less understood. We discovered that the mitochondrial metabolic enzyme hexokinase 2 (HK2) localizes to the nucleus in leukaemic and normal haematopoietic stem cells. Overexpression of nuclear HK2 increases leukaemic stem cell properties and decreases differentiation, whereas selective nuclear HK2 knockdown promotes differentiation and decreases stem cell function. Nuclear HK2 localization is phosphorylation-dependent, requires active import and export, and regulates differentiation independently of its enzymatic activity. HK2 interacts with nuclear proteins regulating chromatin openness, increasing chromatin accessibilities at leukaemic stem cell-positive signature and DNA-repair sites. Nuclear HK2 overexpression decreases double-strand breaks and confers chemoresistance, which may contribute to the mechanism by which leukaemic stem cells resist DNA-damaging agents. Thus, we describe a non-canonical mechanism by which mitochondrial enzymes influence stem cell function independently of their metabolic function.
© 2022. The Author(s).
Conflict of interest statement
A.D.S. has received research funding from Takeda Pharmaceuticals and Medivir AB as well as consulting fees/honorarium from Takeda, Novartis, Jazz and Otsuka Pharmaceuticals. A.D.S. is named on a patent application for the use of DNT cells to treat AML (US patent application no. US62/971,534). The remaining authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

