A novel non-invasive method allowing for discovery of pathologically relevant proteins from small airways
- PMID: 35668386
- PMCID: PMC9167914
- DOI: 10.1186/s12014-022-09348-y
A novel non-invasive method allowing for discovery of pathologically relevant proteins from small airways
Erratum in
-
Correction to: A novel non‑invasive method allowing for discovery of pathologically relevant proteins from small airways.Clin Proteomics. 2022 Jul 18;19(1):29. doi: 10.1186/s12014-022-09363-z. Clin Proteomics. 2022. PMID: 35843940 Free PMC article. No abstract available.
Abstract
Background: There is a lack of early and precise biomarkers for personalized respiratory medicine. Breath contains an aerosol of droplet particles, which are formed from the epithelial lining fluid when the small airways close and re-open during inhalation succeeding a full expiration. These particles can be collected by impaction using the PExA® method (Particles in Exhaled Air), and are derived from an area of high clinical interest previously difficult to access, making them a potential source of biomarkers reflecting pathological processes in the small airways.
Research question: Our aim was to investigate if PExA method is useful for discovery of biomarkers that reflect pathology of small airways.
Methods and analysis: Ten healthy controls and 20 subjects with asthma, of whom 10 with small airway involvement as indicated by a high lung clearance index (LCI ≥ 2.9 z-score), were examined in a cross-sectional design, using the PExA instrument. The samples were analysed with the SOMAscan proteomics platform (SomaLogic Inc.).
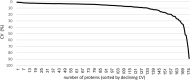
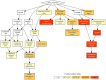
Results: Two hundred-seven proteins were detected in up to 80% of the samples. Nine proteins showed differential abundance in subjects with asthma and high LCI as compared to healthy controls. Two of these were less abundant (ALDOA4, C4), and seven more abundant (FIGF, SERPINA1, CD93, CCL18, F10, IgM, IL1RAP). sRAGE levels were lower in ex-smokers (n = 14) than in never smokers (n = 16). Gene Ontology (GO) annotation database analyses revealed that the PEx proteome is enriched in extracellular proteins associated with extracellular exosome-vesicles and innate immunity.
Conclusion: The applied analytical method was reproducible and allowed identification of pathologically interesting proteins in PEx samples from asthmatic subjects with high LCI. The results suggest that PEx based proteomics is a novel and promising approach to study respiratory diseases with small airway involvement.
Keywords: Asthma; Biomarker; Breath; Exhaled air; Non-invasive; Non-volatiles; Precision medicine; Proteomics; Small airways.
© 2022. The Author(s).
Conflict of interest statement
Anna-Carin Olin is reporting competing interests as she is one of the inventors of the PExA method, and boardmember and chairholder of PExA AB. Emilia Viklund is reporting a minor chairhold in PExA AB. Dr Östling reports personal fees from PExA AB during the conduct of the study; and Employed by PExA AB while writing the manuscript but not during the planning and completion of the study.
Figures


References
-
- Postma DS, Brightling C, Baldi S, Van den Berge M, Fabbri LM, Gagnatelli A, Papi A, Van der Molen T, Rabe KF, Siddiqui S, Singh D, Nicolini G, Kraft M, Group AS Exploring the relevance and extent of small airways dysfunction in asthma (ATLANTIS): baseline data from a prospective cohort study. Lancet Respir Med. 2019;7(5):402–416. doi: 10.1016/S2213-2600(19)30049-9. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
