Low-cost sample preservation methods for high-throughput processing of rumen microbiomes
- PMID: 35668514
- PMCID: PMC9171989
- DOI: 10.1186/s42523-022-00190-z
Low-cost sample preservation methods for high-throughput processing of rumen microbiomes
Abstract
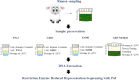
Background: The use of rumen microbial community (RMC) profiles to predict methane emissions has driven interest in ruminal DNA preservation and extraction protocols that can be processed cheaply while also maintaining or improving DNA quality for RMC profiling. Our standard approach for preserving rumen samples, as defined in the Global Rumen Census (GRC), requires time-consuming pre-processing steps of freeze drying and grinding prior to international transportation and DNA extraction. This impedes researchers unable to access sufficient funding or infrastructure. To circumvent these pre-processing steps, we investigated three methods of preserving rumen samples for subsequent DNA extraction, based on existing lysis buffers Tris-NaCl-EDTA-SDS (TNx2) and guanidine hydrochloride (GHx2), or 100% ethanol.
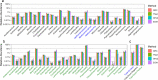
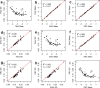
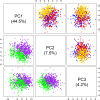
Results: Rumen samples were collected via stomach intubation from 151 sheep at two time-points 2 weeks apart. Each sample was separated into four subsamples and preserved using the three preservation methods and the GRC method (n = 4 × 302). DNA was extracted and sequenced using Restriction Enzyme-Reduced Representation Sequencing to generate RMC profiles. Differences in DNA yield, quality and integrity, and sequencing metrics were observed across the methods (p < 0.0001). Ethanol exhibited poorer quality DNA (A260/A230 < 2) and more failed samples compared to the other methods. Samples preserved using the GRC method had smaller relative abundances in gram-negative genera Anaerovibrio, Bacteroides, Prevotella, Selenomonas, and Succiniclasticum, but larger relative abundances in the majority of 56 additional genera compared to TNx2 and GHx2. However, log10 relative abundances across all genera and time-points for TNx2 and GHx2 were on average consistent (R2 > 0.99) but slightly more variable compared to the GRC method. Relative abundances were moderately to highly correlated (0.68 ± 0.13) between methods for samples collected within a time-point, which was greater than the average correlation (0.17 ± 0.11) between time-points within a preservation method.
Conclusions: The two modified lysis buffers solutions (TNx2 and GHx2) proposed in this study were shown to be viable alternatives to the GRC method for RMC profiling in sheep. Use of these preservative solutions reduces cost and improves throughput associated with processing and sequencing ruminal samples. This development could significantly advance implementation of RMC profiles as a tool for breeding ruminant livestock.
Keywords: Genotyping-by-sequencing; PstI; RE-RRS; Rumen microbial profiles; Rumen microbiology; Superorganism.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Huws SA, Creevey CJ, Oyama LB, Mizrahi I, Denman SE, Popova M, Muñoz-Tamayo R, Forano E, Waters SM, Hess M. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: past, present, and future. Front Microbiol. 2018;9:2161. doi: 10.3389/fmicb.2018.02161. - DOI - PMC - PubMed
Grants and funding
- PhD scholarship/Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES)
- SOW14-AGR-GPLER-SP5-SR/Global Research Alliance
- SOW14-AGR-GPLER-SP5-SR/Global Research Alliance
- C10X1807/Ministry of Business, Innovation and Employment
- C10X1807/Ministry of Business, Innovation and Employment
LinkOut - more resources
Full Text Sources
