RNA allows identifying the consumption of carrion prey
- PMID: 35668675
- PMCID: PMC9541938
- DOI: 10.1111/1755-0998.13659
RNA allows identifying the consumption of carrion prey
Abstract
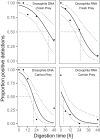
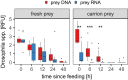
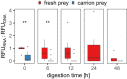
Facultative scavenging by predatory carnivores is a prevalent but frequently underestimated feeding strategy. DNA-based methods for diet analysis, however, do not allow to distinguish between scavenging and predation, thus, the significance of scavenging on population dynamics and resource partitioning is widely unknown. Here, we present a methodological innovation to differentiate between scavenging and fresh prey consumption using prey RNA as a target molecule. We hypothesized that the rapid post-mortem breakdown of RNA in prey tissue should lead to a significantly lower detection probability of prey RNA than DNA when carrion rather than fresh prey is consumed. To test this hypothesis, ground beetles (Pseudoophonus rufipes [De Geer]) were offered either fresh or 1-day-old dead Drosophila melanogaster fruit flies (carrion). The detectability of prey RNA and DNA in the beetles' regurgitates was assessed with diagnostic Drosophila-specific RT-PCR and PCR assays at 0, 6, 12, 24 and 48 h post-feeding. After fresh fly consumption, prey RNA and DNA were detectable equally well at all times. When carrion prey was consumed, the detection strength of prey RNA immediately after feeding was significantly lower than that of prey DNA and reached zero in most samples within 6 h of digestion. Our findings provide evidence that prey RNA allows distinguishing between the consumption of fresh and scavenged prey, thereby overcoming a long-known weakness of molecular diet analysis. The assessment of prey RNA offers a generally applicable approach for examining the importance of scavenging in food webs to unravel its functional consequences for populations, communities, and ecosystems.
Keywords: food webs; molecular diet analysis; scavenging; top-down control; trophic cascades; trophic linking.
© 2022 The Authors. Molecular Ecology Resources published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Detecting predation and scavenging by DNA gut-content analysis: a case study using a soil insect predator-prey system.Oecologia. 2005 Jan;142(3):344-52. doi: 10.1007/s00442-004-1736-7. Epub 2004 Oct 29. Oecologia. 2005. PMID: 15517409
-
Scavenging: how carnivores and carrion structure communities.Trends Ecol Evol. 2011 Mar;26(3):129-35. doi: 10.1016/j.tree.2010.12.011. Epub 2011 Feb 2. Trends Ecol Evol. 2011. PMID: 21295371
-
The significance of facultative scavenging in generalist predator nutrition: detecting decayed prey in the guts of predators using PCR.Mol Ecol. 2005 Nov;14(13):4147-58. doi: 10.1111/j.1365-294X.2005.02732.x. Mol Ecol. 2005. PMID: 16262865
-
Inter-specific interactions linking predation and scavenging in terrestrial vertebrate assemblages.Biol Rev Camb Philos Soc. 2014 Nov;89(4):1042-54. doi: 10.1111/brv.12097. Epub 2014 Mar 7. Biol Rev Camb Philos Soc. 2014. PMID: 24602047 Review.
-
Enemies with benefits: integrating positive and negative interactions among terrestrial carnivores.Ecol Lett. 2020 May;23(5):902-918. doi: 10.1111/ele.13489. Epub 2020 Mar 17. Ecol Lett. 2020. PMID: 32185877 Review.
Cited by
-
Molecular diet analysis enables detection of diatom and cyanobacteria DNA in the gut of Macoma balthica.PLoS One. 2022 Nov 23;17(11):e0278070. doi: 10.1371/journal.pone.0278070. eCollection 2022. PLoS One. 2022. PMID: 36417463 Free PMC article.
-
Utilizing the state of environmental DNA (eDNA) to incorporate time-scale information into eDNA analysis.Proc Biol Sci. 2023 May 31;290(1999):20230979. doi: 10.1098/rspb.2023.0979. Epub 2023 May 31. Proc Biol Sci. 2023. PMID: 37253423 Free PMC article. Review.
References
-
- Beasley, J. C. , Olson, Z. H. , & DeVault, T. L. (2012). Carrion cycling in food webs: Comparisons among terrestrial and marine ecosystems. Oikos, 121(7), 1021–1026. 10.1111/j.1600-0706.2012.20353.x - DOI
-
- Boecklen, W. J. , Yarnes, C. T. , Cook, B. A. , & James, A. C. (2011). On the use of stable isotopes in trophic ecology. Annual Review of Ecology, Evolution, and Systematics, 42(42), 411–440. 10.1146/annurev-ecolsys-102209-144726 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

