Tea Ingredients Have Anti-coronavirus Disease 2019 (COVID-19) Targets Based on Bioinformatics Analyses and Pharmacological Effects on LPS-Stimulated Macrophages
- PMID: 35669076
- PMCID: PMC9163550
- DOI: 10.3389/fnut.2022.875765
Tea Ingredients Have Anti-coronavirus Disease 2019 (COVID-19) Targets Based on Bioinformatics Analyses and Pharmacological Effects on LPS-Stimulated Macrophages
Abstract
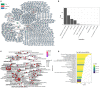
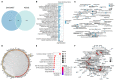
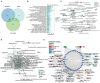
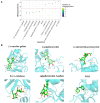
Coronavirus disease 2019 (COVID-19) is a contagious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that caused millions of deaths and lacks treatment. Although several studies have focused on the major component of green tea, epigallocatechin 3-gallate (EGCG), which is efficient in preventing COVID-19, systemic analyses of the anti-COVID-19 potential of green tea remain insufficient. Here, we co-analyzed the target genes of tea ingredients and COVID-19 signature genes and found that epigallocatechin 3-acetalbehyde was capable of reversing the major molecular processes of COVID-19 (MAPK and NF-κB activation). These findings were further supported by Western blotting (WB), immunofluorescence, and quantitative polymerase chain reaction (qPCR) in LPS-stimulated macrophages. Moreover, using molecular docking analysis, we identified three tea ingredients ((-)-catechin gallate, D-(+)-cellobiose, and EGCG) that may interact with the vital SARS-CoV-2 protein, 5R84, compared with the qualified 5R84 ligand WGS. Thus, our results indicated that tea ingredients have the potential to treat COVID-19 by suppressing the COVID-19 signature genes and interacting with the vital SARS-CoV-2 protein.
Keywords: COVID-19; key targets; macrophage; molecular docking; network pharmacology; tea ingredients.
Copyright © 2022 Wang, Tao, Wang, Shi, Yan, Zhang, Sun and Yao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Integrated Network Pharmacology and Molecular Docking to Elucidate the Efficacy and Potential Mechanisms of Tea Ingredients in Sepsis Treatment.Biochem Genet. 2024 Jun;62(3):2253-2267. doi: 10.1007/s10528-023-10530-6. Epub 2023 Oct 30. Biochem Genet. 2024. PMID: 37902912
-
Therapeutic potential of green tea catechin, (-)-epigallocatechin-3-O-gallate (EGCG) in SARS-CoV-2 infection: Major interactions with host/virus proteases.Phytomed Plus. 2023 Feb;3(1):100402. doi: 10.1016/j.phyplu.2022.100402. Epub 2022 Dec 30. Phytomed Plus. 2023. PMID: 36597465 Free PMC article. Review.
-
The green tea catechin epigallocatechin gallate inhibits SARS-CoV-2 infection.J Gen Virol. 2021 Apr;102(4):001574. doi: 10.1099/jgv.0.001574. J Gen Virol. 2021. PMID: 33830908 Free PMC article.
-
A molecular docking study of EGCG and theaflavin digallate with the druggable targets of SARS-CoV-2.Comput Biol Med. 2021 Feb;129:104137. doi: 10.1016/j.compbiomed.2020.104137. Epub 2020 Nov 23. Comput Biol Med. 2021. PMID: 33302163 Free PMC article.
-
EGCG, a Green Tea Catechin, as a Potential Therapeutic Agent for Symptomatic and Asymptomatic SARS-CoV-2 Infection.Molecules. 2021 Feb 24;26(5):1200. doi: 10.3390/molecules26051200. Molecules. 2021. PMID: 33668085 Free PMC article. Review.
Cited by
-
Causal associations of tea intake with COVID-19 infection and severity.Front Nutr. 2023 Jan 4;9:1005466. doi: 10.3389/fnut.2022.1005466. eCollection 2022. Front Nutr. 2023. PMID: 36687732 Free PMC article.
References
-
- Khandia R, Singhal S, Alqahtani T, Kamal MA, El-Shall NA. F. Nainu, et al. Emergence of SARS-CoV-2 Omicron (B11529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ Res. (2022) 209:112816. 10.1016/j.envres.2022.112816 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

