Effect of CRISPR Knockout of AXIN1 or ARID1A on Proliferation and Migration of Porcine Hepatocellular Carcinoma
- PMID: 35669430
- PMCID: PMC9163418
- DOI: 10.3389/fonc.2022.904031
Effect of CRISPR Knockout of AXIN1 or ARID1A on Proliferation and Migration of Porcine Hepatocellular Carcinoma
Abstract
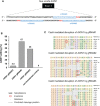
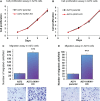
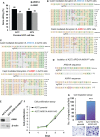
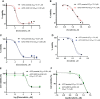
Hepatocellular carcinoma (HCC) is an aggressive disease lacking effective treatment. Animal models of HCC are necessary for preclinical evaluation of the safety and efficacy of novel therapeutics. Large animal models of HCC allow testing image-guided locoregional therapies, which are widely used in the management of HCC. Models with precise tumor mutations mimicking human HCC provide valuable tools for testing precision medicine. AXIN1 and ARID1A are two of the most frequently mutated genes in human HCC. Here, we investigated the effects of knockout of AXIN1 and/or ARID1A on proliferation, migration, and chemotherapeutic susceptibility of porcine HCC cells and we developed subcutaneous tumors harboring these mutations in pigs. Gene knockout was achieved by CRISPR/Cas9 and was validated by Next Generation Sequencing. AXIN1 knockout increased the migration of porcine HCC cells but did not alter the cell proliferation. Knockout of ARID1A increased both the proliferation and migration of porcine HCC cells. Simultaneous knockout of AXIN1 and ARID1A increased the migration, but did not alter the proliferation of porcine HCC cells. The effect of gene knockout on the response of porcine HCC cells to two of the most commonly used systemic and locoregional HCC treatments was investigated; sorafenib and doxorubicin, respectively. Knockout of AXIN1 and/or ARID1A did not alter the susceptibility of porcine HCC cells to sorafenib or doxorubicin. Autologous injection of CRISPR edited HCC cells resulted in development of subcutaneous tumors in pigs, which harbored the anticipated edits in AXIN1 and/or ARID1A. This study elucidates the effects of CRISPR-mediated knockout of HCC-associated genes in porcine HCC cells, and lays the foundation for development and utilization of genetically-tailored porcine HCC models for in vivo testing of novel therapeutic approaches in a clinically-relevant large animal model.
Keywords: ARID1A; AXIN1; CRISPR; gene knockout; hepatocellular carcinoma; liver cancer; porcine models.
Copyright © 2022 Elkhadragy, Dasteh Goli, Totura, Carlino, Regan, Guzman, Schook, Gaba and Schachtschneider.
Conflict of interest statement
LS, RG, and KS have received research support from the United States Department of Defense, the United States National Institutes of Health, Guerbet USA LLC, Janssen Research & Development LLC, NeoTherma Oncology, and TriSalus Life Sciences, and are scientific consultants for Sus Clinicals Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
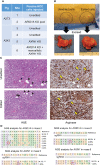
Figures



References
LinkOut - more resources
Full Text Sources

