ZNF384: A Potential Therapeutic Target for Psoriasis and Alzheimer's Disease Through Inflammation and Metabolism
- PMID: 35669784
- PMCID: PMC9163351
- DOI: 10.3389/fimmu.2022.892368
ZNF384: A Potential Therapeutic Target for Psoriasis and Alzheimer's Disease Through Inflammation and Metabolism
Abstract
Background: Psoriasis is an immune-related skin disease notable for its chronic inflammation of the entire system. Alzheimer's disease (AD) is more prevalent in psoriasis than in the general population. Immune-mediated pathophysiologic processes may link these two diseases, but the mechanism is still unclear. This article aimed to explore potential molecular mechanisms in psoriasis and AD.
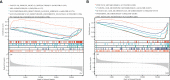
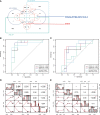
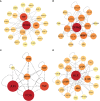
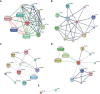
Methods: Gene expression profiling data of psoriasis and AD were acquired in the Gene Expression Omnibus (GEO) database. Gene Set Enrichment Analysis (GSEA) and single-sample GSEA (ssGSEA) were first applied in two datasets. Differentially expressed genes (DEGs) of two diseases were identified, and common DEGs were selected. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed to explore common biological pathways. Signature transcription factors (STFs) were identified and their diagnostic values was calculated by receiver operating characteristic (ROC) curve analysis in the exploration cohort and verified in the validation cohort. The expression levels of STFs were further investigated in the validation cohort and the GTEx Portal Database. Additionally, four kinds of interaction analysis were performed: correlation analysis among STFs, gene-gene, chemical-protein, and protein-ligand interaction analyses. In the end, we predicted the transcription factor that potentially regulates STFs.
Results: Biosynthesis and metabolic pathways were enriched in GSEA analysis. In ssGSEA analysis, most immunoreaction gene lists exhibited differential enrichment in psoriasis cases, whereas three receptor-related gene lists did in AD. The KEGG analysis of common DEGs redetermined inflammatory and metabolic pathways essential in both diseases. 5 STFs (PPARG, ZFPM2, ZNF415, HLX, and ANHX) were screened from common DEGs. The ROC analysis indicated that all STFs have diagnostic values in two diseases, especially ZFPM2. The correlation analysis, gene-gene, chemical-protein, and protein-ligand interaction analyses suggested that STFs interplay and involve inflammation and aberrant metabolism. Eventually, ZNF384 was the predicted transcription factor regulating PPARG, ZNF415, HLX, and ANHX.
Conclusions: The STFs (PPARG, ZFPM2, ZNF415, HLX, and ANHX) may increase the morbidity rate of AD in psoriasis by initiating a positive feedback loop of excessive inflammation and metabolic disorders. ZNF384 is a potential therapeutic target for psoriasis and AD by regulating PPARG, ZNF415, HLX, and ANHX.
Keywords: Alzheimer’s disease; inflammation; metabolic disorders; psoriasis; transcription factors.
Copyright © 2022 Liu, Yuan, Su, Liu, Zhuang and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

