Multi-omic analysis shows REVEILLE clock genes are involved in carbohydrate metabolism and proteasome function
- PMID: 35670757
- PMCID: PMC9516735
- DOI: 10.1093/plphys/kiac269
Multi-omic analysis shows REVEILLE clock genes are involved in carbohydrate metabolism and proteasome function
Abstract
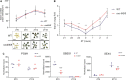
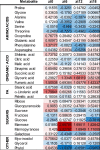
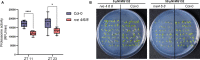
Plants are able to sense changes in their light environments, such as the onset of day and night, as well as anticipate these changes in order to adapt and survive. Central to this ability is the plant circadian clock, a molecular circuit that precisely orchestrates plant cell processes over the course of a day. REVEILLE (RVE) proteins are recently discovered members of the plant circadian circuitry that activate the evening complex and PSEUDO-RESPONSE REGULATOR genes to maintain regular circadian oscillation. The RVE8 protein and its two homologs, RVE 4 and 6 in Arabidopsis (Arabidopsis thaliana), have been shown to limit the length of the circadian period, with rve 4 6 8 triple-knockout plants possessing an elongated period along with increased leaf surface area, biomass, cell size, and delayed flowering relative to wild-type Col-0 plants. Here, using a multi-omics approach consisting of phenomics, transcriptomics, proteomics, and metabolomics we draw new connections between RVE8-like proteins and a number of core plant cell processes. In particular, we reveal that loss of RVE8-like proteins results in altered carbohydrate, organic acid, and lipid metabolism, including a starch excess phenotype at dawn. We further demonstrate that rve 4 6 8 plants have lower levels of 20S proteasome subunits and possess significantly reduced proteasome activity, potentially explaining the increase in cell-size observed in RVE8-like mutants. Overall, this robust, multi-omic dataset provides substantial insight into the far-reaching impact RVE8-like proteins have on the diel plant cell environment.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




Similar articles
-
COLD REGULATED GENE 27 and 28 antagonize the transcriptional activity of the RVE8/LNK1/LNK2 circadian complex.Plant Physiol. 2023 Jul 3;192(3):2436-2456. doi: 10.1093/plphys/kiad210. Plant Physiol. 2023. PMID: 37017001 Free PMC article.
-
The circadian clock mutant lhy cca1 elf3 paces starch mobilization to dawn despite severely disrupted circadian clock function.Plant Physiol. 2022 Aug 1;189(4):2332-2356. doi: 10.1093/plphys/kiac226. Plant Physiol. 2022. PMID: 35567528 Free PMC article.
-
REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock.PLoS Genet. 2011 Mar;7(3):e1001350. doi: 10.1371/journal.pgen.1001350. Epub 2011 Mar 31. PLoS Genet. 2011. PMID: 21483796 Free PMC article.
-
Chemical biology to dissect molecular mechanisms underlying plant circadian clocks.New Phytol. 2022 Aug;235(4):1336-1343. doi: 10.1111/nph.18298. Epub 2022 Jun 23. New Phytol. 2022. PMID: 35661165 Review.
-
Illuminating the Arabidopsis circadian epigenome: Dynamics of histone acetylation and deacetylation.Curr Opin Plant Biol. 2022 Oct;69:102268. doi: 10.1016/j.pbi.2022.102268. Epub 2022 Jul 31. Curr Opin Plant Biol. 2022. PMID: 35921796 Review.
Cited by
-
Transcriptional activation and repression in the plant circadian clock: Revisiting core oscillator feedback loops and output pathways.Plant Commun. 2025 Aug 11;6(8):101415. doi: 10.1016/j.xplc.2025.101415. Epub 2025 Jun 10. Plant Commun. 2025. PMID: 40500965 Free PMC article. Review.
-
Systems-level proteomics and metabolomics reveals the diel molecular landscape of diverse kale cultivars.Front Plant Sci. 2023 Jul 28;14:1170448. doi: 10.3389/fpls.2023.1170448. eCollection 2023. Front Plant Sci. 2023. PMID: 37575922 Free PMC article.
-
Focus on circadian rhythms.Plant Physiol. 2022 Sep 28;190(2):921-923. doi: 10.1093/plphys/kiac353. Plant Physiol. 2022. PMID: 35900174 Free PMC article. No abstract available.
-
Circadian regulation of metabolism across photosynthetic organisms.Plant J. 2023 Nov;116(3):650-668. doi: 10.1111/tpj.16405. Epub 2023 Aug 2. Plant J. 2023. PMID: 37531328 Free PMC article. Review.
-
Computational Reconstruction of the Transcription Factor Regulatory Network Induced by Auxin in Arabidopsis thaliana L.Plants (Basel). 2024 Jul 10;13(14):1905. doi: 10.3390/plants13141905. Plants (Basel). 2024. PMID: 39065433 Free PMC article.
References
-
- Aarabi F, Naake T, Fernie AR, Hoefgen R (2020) Coordinating sulfur pools under sulfate deprivation. Trends Plant Sci 25: 1227–1239 - PubMed
-
- Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with imageJ. Biophoton Int 11: 36–42
-
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR (2003) A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 6: 251–257 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

