Phylotranscriptomic analyses reveal multiple whole-genome duplication events, the history of diversification and adaptations in the Araceae
- PMID: 35671385
- PMCID: PMC9904356
- DOI: 10.1093/aob/mcac062
Phylotranscriptomic analyses reveal multiple whole-genome duplication events, the history of diversification and adaptations in the Araceae
Abstract
Background and aims: The Araceae are one of the most diverse monocot families with numerous morphological and ecological novelties. Plastid and mitochondrial genes have been used to investigate the phylogeny and to interpret shifts in the pollination biology and biogeography of the Araceae. In contrast, the role of whole-genome duplication (WGD) in the evolution of eight subfamilies remains unclear.
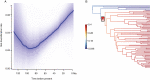
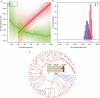
Methods: New transcriptomes or low-depth whole-genome sequences of 65 species were generated through Illumina sequencing. We reconstructed the phylogenetic relationships of Araceae using concatenated and species tree methods, and then estimated the age of major clades using TreePL. We inferred the WGD events by Ks and gene tree methods. We investigated the diversification patterns applying time-dependent and trait-dependent models. The expansions of gene families and functional enrichments were analysed using CAFE and InterProScan.
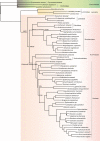
Key results: Gymnostachydoideae was the earliest diverging lineage followed successively by Orontioideae, Lemnoideae and Lasioideae. In turn, they were followed by the clade of 'bisexual climbers' comprised of Pothoideae and Monsteroideae, which was resolved as the sister to the unisexual flowers clade of Zamioculcadoideae and Aroideae. A special WGD event ψ (psi) shared by the True-Araceae clade occurred in the Early Cretaceous. Net diversification rates first declined and then increased through time in the Araceae. The best diversification rate shift along the stem lineage of the True-Araceae clade was detected, and net diversification rates were enhanced following the ψ-WGD. Functional enrichment analyses revealed that some genes, such as those encoding heat shock proteins, glycosyl hydrolase and cytochrome P450, expanded within the True-Araceae clade.
Conclusions: Our results improve our understanding of aroid phylogeny using the large number of single-/low-copy nuclear genes. In contrast to the Proto-Araceae group and the lemnoid clade adaption to aquatic environments, our analyses of WGD, diversification and functional enrichment indicated that WGD may play a more important role in the evolution of adaptations to tropical, terrestrial environments in the True-Araceae clade. These insights provide us with new resources to interpret the evolution of the Araceae.
Keywords: Araceae; WGDs; adaptation; diversification; phylogenomics.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Intergeneric and infrafamilial phylogeny of subfamily Monsteroideae (Araceae) revealed by chloroplast trnL-F sequences.Am J Bot. 2004 Mar;91(3):490-8. doi: 10.3732/ajb.91.3.490. Am J Bot. 2004. PMID: 21653404
-
Phylogenetic relationships of aroids and duckweeds (Araceae) inferred from coding and noncoding plastid DNA.Am J Bot. 2008 Sep;95(9):1153-65. doi: 10.3732/ajb.0800073. Am J Bot. 2008. PMID: 21632433
-
Impact of whole-genome duplication events on diversification rates in angiosperms.Am J Bot. 2018 Mar;105(3):348-363. doi: 10.1002/ajb2.1060. Epub 2018 May 2. Am J Bot. 2018. PMID: 29719043
-
Whole-genome duplication in teleost fishes and its evolutionary consequences.Mol Genet Genomics. 2014 Dec;289(6):1045-60. doi: 10.1007/s00438-014-0889-2. Epub 2014 Aug 5. Mol Genet Genomics. 2014. PMID: 25092473 Review.
-
Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model.Curr Opin Plant Biol. 2012 Apr;15(2):147-53. doi: 10.1016/j.pbi.2012.03.011. Epub 2012 Apr 3. Curr Opin Plant Biol. 2012. PMID: 22480429 Review.
Cited by
-
A plastid phylogenomic framework for the palm family (Arecaceae).BMC Biol. 2023 Mar 8;21(1):50. doi: 10.1186/s12915-023-01544-y. BMC Biol. 2023. PMID: 36882831 Free PMC article.
-
A roadmap of phylogenomic methods for studying polyploid plant genera.Appl Plant Sci. 2024 Apr 22;12(4):e11580. doi: 10.1002/aps3.11580. eCollection 2024 Jul-Aug. Appl Plant Sci. 2024. PMID: 39184196 Free PMC article.
-
Polyploidy: its consequences and enabling role in plant diversification and evolution.Ann Bot. 2023 Feb 7;131(1):1-10. doi: 10.1093/aob/mcac132. Ann Bot. 2023. PMID: 36282971 Free PMC article.
-
Evolutionary Dynamics of Chromatin Structure and Duplicate Gene Expression in Diploid and Allopolyploid Cotton.Mol Biol Evol. 2024 May 3;41(5):msae095. doi: 10.1093/molbev/msae095. Mol Biol Evol. 2024. PMID: 38758089 Free PMC article.
-
Evolution of the Colocasiomyia gigantea Species Group (Diptera: Drosophilidae): Phylogeny, Biogeography and Shift of Host Use.Insects. 2022 Jul 18;13(7):647. doi: 10.3390/insects13070647. Insects. 2022. PMID: 35886823 Free PMC article.
References
-
- Arrigo N, Barker MS. 2012. Rarely successful polyploids and their legacy in plant genomes. Current Opinion in Plant Biology 15: 140–146. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

