A novel mRNA decay inhibitor abolishes pathophysiological cellular transition
- PMID: 35672286
- PMCID: PMC9174231
- DOI: 10.1038/s41420-022-01076-4
A novel mRNA decay inhibitor abolishes pathophysiological cellular transition
Abstract
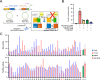
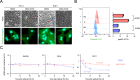
In cells, mRNA synthesis and decay are influenced by each other, and their balance is altered by either external or internal cues, resulting in changes in cell dynamics. We previously reported that it is important that an array of mRNAs that shape a phenotype are degraded before cellular transitions, such as cellular reprogramming and differentiation. In adipogenesis, the interaction between DDX6 and 4E-T had a definitive impact on the pathway in the processing body (PB). We screened a library of α-helix analogs with an alkaloid-like backbone to identify compounds that inhibit the binding between DDX6 and 4E-T proteins, which occurs between the α-helix of structured and internally disordered proteins. IAMC-00192 was identified as a lead compound. This compound directly inhibited the interaction between DDX6 and 4E-T. IAMC-00192 inhibited the temporal increase in PB formation that occurs during adipogenesis and epithelial-mesenchymal transition (EMT) and significantly suppressed these cellular transitions. In the EMT model, the half-life of preexisting mRNAs in PBs was extended twofold by the compound. The novel inhibitor of RNA decay not only represents a potentially useful tool to analyze in detail the pathological conditions affected by RNA decay and how it regulates the pathological state. The identification of this inhibitor may lead to the discovery of a first-in-class RNA decay inhibitor drug.
© 2022. The Author(s).
Conflict of interest statement
H.K. is the Chief Executive Officer of Oita University Institute of Advanced Medicine, Inc., which provided the small molecule library. Other co-authors have no conflicts of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

