JMJD3 intrinsically disordered region links the 3D-genome structure to TGFβ-dependent transcription activation
- PMID: 35672304
- PMCID: PMC9174158
- DOI: 10.1038/s41467-022-30614-y
JMJD3 intrinsically disordered region links the 3D-genome structure to TGFβ-dependent transcription activation
Abstract
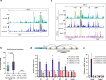
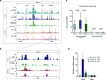
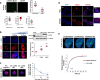
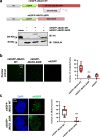
Enhancers are key regulatory elements that govern gene expression programs in response to developmental signals. However, how multiple enhancers arrange in the 3D-space to control the activation of a specific promoter remains unclear. To address this question, we exploited our previously characterized TGFβ-response model, the neural stem cells, focusing on a ~374 kb locus where enhancers abound. Our 4C-seq experiments reveal that the TGFβ pathway drives the assembly of an enhancer-cluster and precise gene activation. We discover that the TGFβ pathway coactivator JMJD3 is essential to maintain these structures. Using live-cell imaging techniques, we demonstrate that an intrinsically disordered region contained in JMJD3 is involved in the formation of phase-separated biomolecular condensates, which are found in the enhancer-cluster. Overall, in this work we uncover novel functions for the coactivator JMJD3, and we shed light on the relationships between the 3D-conformation of the chromatin and the TGFβ-driven response during mammalian neurogenesis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

