Linked-read sequencing for detecting short tandem repeat expansions
- PMID: 35672336
- PMCID: PMC9174224
- DOI: 10.1038/s41598-022-13024-4
Linked-read sequencing for detecting short tandem repeat expansions
Abstract
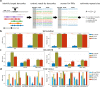
Detection of short tandem repeat (STR) expansions with standard short-read sequencing is challenging due to the difficulty in mapping multicopy repeat sequences. In this study, we explored how the long-range sequence information of barcode linked-read sequencing (BLRS) can be leveraged to improve repeat-read detection. We also devised a novel algorithm using BLRS barcodes for distance estimation and evaluated its application for STR genotyping. Both approaches were designed for genotyping large expansions (> 1 kb) that cannot be sized accurately by existing methods. Using simulated and experimental data of genomes with STR expansions from multiple BLRS platforms, we validated the utility of barcode and phasing information in attaining better STR genotypes compared to standard short-read sequencing. Although the coverage bias of extremely GC-rich STRs is an important limitation of BLRS, BLRS is an effective strategy for genotyping many other STR loci.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Wang O, Chin R, Cheng X, Wu M, Mao Q, Tang J, et al. Efficient and unique co-barcoding of second-generation sequencing reads from long DNA molecules enabling cost effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res. 2019;29(5):798–808. doi: 10.1101/gr.245126.118. - DOI - PMC - PubMed
-
- Chen Z, Pham L, Wu T-C, Mo G, Xia Y, Chang PL, et al. Ultralow-input single-tube linked-read library method enables short-read second-generation sequencing systems to routinely generate highly accurate and economical long-range sequencing information. Genome Res. 2020;30:898–909. doi: 10.1101/gr.260380.119. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

