Cardiac fibroblasts regulate the development of heart failure via Htra3-TGF-β-IGFBP7 axis
- PMID: 35672400
- PMCID: PMC9174232
- DOI: 10.1038/s41467-022-30630-y
Cardiac fibroblasts regulate the development of heart failure via Htra3-TGF-β-IGFBP7 axis
Abstract
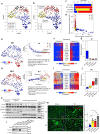
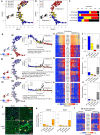
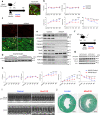
Tissue fibrosis and organ dysfunction are hallmarks of age-related diseases including heart failure, but it remains elusive whether there is a common pathway to induce both events. Through single-cell RNA-seq, spatial transcriptomics, and genetic perturbation, we elucidate that high-temperature requirement A serine peptidase 3 (Htra3) is a critical regulator of cardiac fibrosis and heart failure by maintaining the identity of quiescent cardiac fibroblasts through degrading transforming growth factor-β (TGF-β). Pressure overload downregulates expression of Htra3 in cardiac fibroblasts and activated TGF-β signaling, which induces not only cardiac fibrosis but also heart failure through DNA damage accumulation and secretory phenotype induction in failing cardiomyocytes. Overexpression of Htra3 in the heart inhibits TGF-β signaling and ameliorates cardiac dysfunction after pressure overload. Htra3-regulated induction of spatio-temporal cardiac fibrosis and cardiomyocyte secretory phenotype are observed specifically in infarct regions after myocardial infarction. Integrative analyses of single-cardiomyocyte transcriptome and plasma proteome in human reveal that IGFBP7, which is a cytokine downstream of TGF-β and secreted from failing cardiomyocytes, is the most predictable marker of advanced heart failure. These findings highlight the roles of cardiac fibroblasts in regulating cardiomyocyte homeostasis and cardiac fibrosis through the Htra3-TGF-β-IGFBP7 pathway, which would be a therapeutic target for heart failure.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- López-Otín C, Kroemer G. Hallmarks of health. Cell. 2021;184:33–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

