An approach to cellular tropism of SARS-CoV-2 through protein-protein interaction and enrichment analysis
- PMID: 35672403
- PMCID: PMC9172986
- DOI: 10.1038/s41598-022-13625-z
An approach to cellular tropism of SARS-CoV-2 through protein-protein interaction and enrichment analysis
Abstract
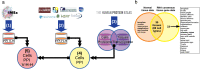
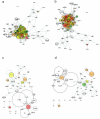
COVID-19, caused by SARS-CoV-2, is a primarily pulmonary disease that can affect several organs, directly or indirectly. To date, there are many questions about the different pathological mechanisms. Here, we generate an approach to identify the cellular-level tropism of SARS-CoV-2 using human proteomics, virus-host interactions, and enrichment analysis. Through a network-based approach, the molecular context was visualized and analyzed. This procedure was also performed for SARS-CoV-1. We obtained proteomes and interactomes from 145 different cells corresponding to 57 different tissues. We discarded the cells without the proteins known for interacting with the virus, such as ACE2 or TMPRSS2. Of the remaining cells, a gradient of susceptibility to infection was observed. In addition, we identified proteins associated with the coagulation cascade that can be directly or indirectly affected by viral proteins. As a whole we identified 55 cells that could be potentially controlled by the virus, with different susceptibilities, mainly being pneumocytes, heart, kidney, liver, or small intestine cells. These results help to explain the molecular context and provide elements for possible treatments in the current situation. This strategy may be useful for other viruses, especially those with limited reported PPI, such as a new virus.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

