Dietary restriction and the transcription factor clock delay eye aging to extend lifespan in Drosophila Melanogaster
- PMID: 35672419
- PMCID: PMC9174495
- DOI: 10.1038/s41467-022-30975-4
Dietary restriction and the transcription factor clock delay eye aging to extend lifespan in Drosophila Melanogaster
Abstract
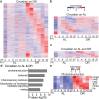
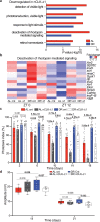
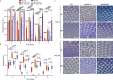
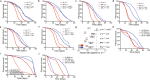
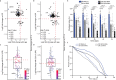
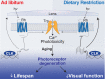
Many vital processes in the eye are under circadian regulation, and circadian dysfunction has emerged as a potential driver of eye aging. Dietary restriction is one of the most robust lifespan-extending therapies and amplifies circadian rhythms with age. Herein, we demonstrate that dietary restriction extends lifespan in Drosophila melanogaster by promoting circadian homeostatic processes that protect the visual system from age- and light-associated damage. Altering the positive limb core molecular clock transcription factor, CLOCK, or CLOCK-output genes, accelerates visual senescence, induces a systemic immune response, and shortens lifespan. Flies subjected to dietary restriction are protected from the lifespan-shortening effects of photoreceptor activation. Inversely, photoreceptor inactivation, achieved via mutating rhodopsin or housing flies in constant darkness, primarily extends the lifespan of flies reared on a high-nutrient diet. Our findings establish the eye as a diet-sensitive modulator of lifespan and indicates that vision is an antagonistically pleiotropic process that contributes to organismal aging.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Daily temperature cycles prolong lifespan and have sex-specific effects on peripheral clock gene expression in Drosophila melanogaster.J Exp Biol. 2021 May 15;224(10):jeb233213. doi: 10.1242/jeb.233213. Epub 2021 May 17. J Exp Biol. 2021. PMID: 33758022
-
Circadian clock genes' overexpression in Drosophila alters diet impact on lifespan.Biogerontology. 2019 Apr;20(2):159-170. doi: 10.1007/s10522-018-9784-2. Epub 2018 Nov 24. Biogerontology. 2019. PMID: 30470951
-
Neuron-specific overexpression of core clock genes improves stress-resistance and extends lifespan of Drosophila melanogaster.Exp Gerontol. 2019 Mar;117:61-71. doi: 10.1016/j.exger.2018.11.005. Epub 2018 Nov 9. Exp Gerontol. 2019. PMID: 30415070
-
Circadian Rhythms and Sleep in Drosophila melanogaster.Genetics. 2017 Apr;205(4):1373-1397. doi: 10.1534/genetics.115.185157. Genetics. 2017. PMID: 28360128 Free PMC article. Review.
-
Circadian regulation of metabolism and healthspan in Drosophila.Free Radic Biol Med. 2018 May 1;119:62-68. doi: 10.1016/j.freeradbiomed.2017.12.025. Epub 2017 Dec 19. Free Radic Biol Med. 2018. PMID: 29277395 Free PMC article. Review.
Cited by
-
New Possibilities for Evaluating the Development of Age-Related Pathologies Using the Dynamical Network Biomarkers Theory.Cells. 2023 Sep 17;12(18):2297. doi: 10.3390/cells12182297. Cells. 2023. PMID: 37759519 Free PMC article. Review.
-
The effect of a pharmaceutical ghrelin agonist on lifespan in C57BL/6J male mice: A controlled experiment.Aging Cell. 2023 Apr;22(4):e13787. doi: 10.1111/acel.13787. Epub 2023 Feb 3. Aging Cell. 2023. PMID: 36734122 Free PMC article.
-
Molecular basis of circadian rhythm divergence between diurnal and nocturnal lepidoperans.iScience. 2025 Mar 12;28(4):112206. doi: 10.1016/j.isci.2025.112206. eCollection 2025 Apr 18. iScience. 2025. PMID: 40212595 Free PMC article.
-
Modelling the ageing dependence of cancer evolutionary trajectories.Nat Rev Cancer. 2025 Jul 10. doi: 10.1038/s41568-025-00838-3. Online ahead of print. Nat Rev Cancer. 2025. PMID: 40640377 Review.
-
Cellular rejuvenation: molecular mechanisms and potential therapeutic interventions for diseases.Signal Transduct Target Ther. 2023 Mar 14;8(1):116. doi: 10.1038/s41392-023-01343-5. Signal Transduct Target Ther. 2023. PMID: 36918530 Free PMC article. Review.
References
-
- Rouyer F. Clock genes: from Drosophila to humans. Bull. Acad. Natl Med. 2015;199:1115–1131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

