Asymmetric host movement reshapes local disease dynamics in metapopulations
- PMID: 35672422
- PMCID: PMC9171740
- DOI: 10.1038/s41598-022-12774-5
Asymmetric host movement reshapes local disease dynamics in metapopulations
Abstract
Understanding how the movement of individuals affects disease dynamics is critical to accurately predicting and responding to the spread of disease in an increasingly interconnected world. In particular, it is not yet known how movement between patches affects local disease dynamics (e.g., whether pathogen prevalence remains steady or oscillates through time). Considering a set of small, archetypal metapopulations, we find three surprisingly simple patterns emerge in local disease dynamics following the introduction of movement between patches: (1) movement between identical patches with cyclical pathogen prevalence dampens oscillations in the destination while increasing synchrony between patches; (2) when patches differ from one another in the absence of movement, adding movement allows dynamics to propagate between patches, alternatively stabilizing or destabilizing dynamics in the destination based on the dynamics at the origin; and (3) it is easier for movement to induce cyclical dynamics than to induce a steady-state. Considering these archetypal networks (and the patterns they exemplify) as building blocks of larger, more realistically complex metapopulations provides an avenue for novel insights into the role of host movement on disease dynamics. Moreover, this work demonstrates a framework for future predictive modelling of disease spread in real populations.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
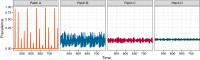
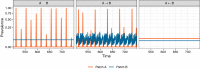
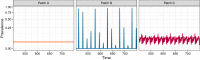


Similar articles
-
Increased migration in host-pathogen metapopulations can cause host extinction.J Theor Biol. 2012 Apr 7;298:1-7. doi: 10.1016/j.jtbi.2011.12.009. Epub 2011 Dec 19. J Theor Biol. 2012. PMID: 22197413
-
Impact of dispersal on the stability of metapopulations.J Theor Biol. 2016 Mar 7;392:1-11. doi: 10.1016/j.jtbi.2015.11.029. Epub 2015 Dec 23. J Theor Biol. 2016. PMID: 26723533
-
From individual movement behaviour to landscape-scale invasion dynamics and management: a case study of lionfish metapopulations.Philos Trans R Soc Lond B Biol Sci. 2019 Sep 16;374(1781):20180057. doi: 10.1098/rstb.2018.0057. Epub 2019 Jul 29. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 31352886 Free PMC article.
-
Infections on the move: how transient phases of host movement influence disease spread.Proc Biol Sci. 2017 Dec 20;284(1869):20171807. doi: 10.1098/rspb.2017.1807. Proc Biol Sci. 2017. PMID: 29263283 Free PMC article. Review.
-
Linking movement behaviour, dispersal and population processes: is individual variation a key?J Anim Ecol. 2009 Sep;78(5):894-906. doi: 10.1111/j.1365-2656.2009.01534.x. Epub 2009 Mar 6. J Anim Ecol. 2009. PMID: 19302396 Review.
Cited by
-
Seasonal contact and migration structure mass epidemics and inform outbreak preparedness in a vulnerable marine mammal.Proc Biol Sci. 2025 Jul;292(2051):20250698. doi: 10.1098/rspb.2025.0698. Epub 2025 Jul 30. Proc Biol Sci. 2025. PMID: 40735844 Free PMC article.
References
-
- Ritchie, H. & Roser, M. Urbanization. Our World in Data (2018). https://ourworldindata.org/urbanization.
-
- Chen, H., Weersink, A., Beaulieu, M., Lee, Y. N. & Nagelschmitz, K. A historical review of changes in farm size in canada. Tech. Rep., University of Guelph, Institute for the Advanced Study of Food and and Agricultural Policy (2019).
-
- Augustin, N., Mugglestone, M. A. & Buckland, S. T. An autologistic model for the spatial distribution of wildlife. J. Appl. Ecol. 339–347 (1996).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

