Cryptic in vitro ubiquitin ligase activity of HDMX towards p53 is probably regulated by an induced fit mechanism
- PMID: 35674210
- PMCID: PMC9254666
- DOI: 10.1042/BSR20220186
Cryptic in vitro ubiquitin ligase activity of HDMX towards p53 is probably regulated by an induced fit mechanism
Abstract
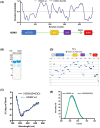
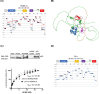
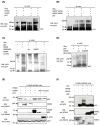
HDMX and its homologue HDM2 are two essential proteins for the cell; after genotoxic stress, both are phosphorylated near to their RING domain, specifically at serine 403 and 395, respectively. Once phosphorylated, both can bind the p53 mRNA and enhance its translation; however, both recognize p53 protein and provoke its degradation under normal conditions. HDM2 has been well-recognized as an E3 ubiquitin ligase, whereas it has been reported that even with the high similarity between the RING domains of the two homologs, HDMX does not have the E3 ligase activity. Despite this, HDMX is needed for the proper p53 poly-ubiquitination. Phosphorylation at serine 395 changes the conformation of HDM2, helping to explain the switch in its activity, but no information on HDMX has been published. Here, we study the conformation of HDMX and its phospho-mimetic mutant S403D, investigate its E3 ligase activity and dissect its binding with p53. We show that phospho-mutation does not change the conformation of the protein, but HDMX is indeed an E3 ubiquitin ligase in vitro; however, in vivo, no activity was found. We speculated that HDMX is regulated by induced fit, being able to switch activity accordingly to the specific partner as p53 protein, p53 mRNA or HDM2. Our results aim to contribute to the elucidation of the contribution of the HDMX to p53 regulation.
Keywords: HDM2; HDMX; Induced fit; MDM2; MDMX; cancer; p53; ubiquitination.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
Allosteric Interactions by p53 mRNA Govern HDM2 E3 Ubiquitin Ligase Specificity under Different Conditions.Mol Cell Biol. 2016 Jul 29;36(16):2195-205. doi: 10.1128/MCB.00113-16. Print 2016 Aug 15. Mol Cell Biol. 2016. PMID: 27215386 Free PMC article.
-
p53 promotes its own polyubiquitination by enhancing the HDM2 and HDMX interaction.Protein Sci. 2018 May;27(5):976-986. doi: 10.1002/pro.3405. Epub 2018 Mar 25. Protein Sci. 2018. PMID: 29524278 Free PMC article.
-
HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53.Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12009-14. doi: 10.1073/pnas.2030930100. Epub 2003 Sep 24. Proc Natl Acad Sci U S A. 2003. PMID: 14507994 Free PMC article.
-
p53 regulation: teamwork between RING domains of Mdm2 and MdmX.Cell Cycle. 2011 Dec 15;10(24):4225-9. doi: 10.4161/cc.10.24.18662. Epub 2011 Dec 15. Cell Cycle. 2011. PMID: 22134240 Review.
-
HDM2 and HDMX Proteins in Human Cancer.Klin Onkol. 2018 Winter;31(Suppl 2):63-70. doi: 10.14735/amko20182S63. Klin Onkol. 2018. PMID: 31023026 Review. English.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

