Cannabis significantly alters DNA methylation of the human ovarian follicle in a concentration-dependent manner
- PMID: 35674367
- PMCID: PMC9247704
- DOI: 10.1093/molehr/gaac022
Cannabis significantly alters DNA methylation of the human ovarian follicle in a concentration-dependent manner
Abstract
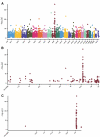
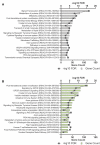
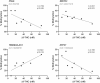
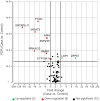
Cannabis is increasingly consumed by women of childbearing age, and the reproductive and epigenetic effects are unknown. The purpose of this study was to evaluate the potential epigenetic implications of cannabis use on the female ovarian follicle. Whole-genome methylation was assessed in granulosa cells from 14 matched case-control patients. Exposure status was determined by liquid chromatography-mass spectrometry (LC-MS/MS) measurements of five cannabis-derived phytocannabinoids in follicular fluid. DNA methylation was measured using the Illumina TruSeq Methyl Capture EPIC kit. Differential methylation, pathway analysis and correlation analysis were performed. We identified 3679 differentially methylated sites, with two-thirds affecting coding genes. A hotspot region on chromosome 9 was associated with two genomic features, a zinc-finger protein (ZFP37) and a long non-coding RNA (FAM225B). There were 2214 differentially methylated genomic features, 19 of which have been previously implicated in cannabis-related epigenetic modifications in other organ systems. Pathway analysis revealed enrichment in G protein-coupled receptor signaling, cellular transport, immune response and proliferation. Applying strict criteria, we identified 71 differentially methylated regions, none of which were previously annotated in this context. Finally, correlation analysis revealed 16 unique genomic features affected by cannabis use in a concentration-dependent manner. Of these, the histone methyltransferases SMYD3 and ZFP37 were hypomethylated, possibly implicating histone modifications as well. Herein, we provide the first DNA methylation profile of human granulosa cells exposed to cannabis. With cannabis increasingly legalized worldwide, further investigation into the heritability and functional consequences of these effects is critical for clinical consultation and for legalization guidelines.
Keywords: DNA methylation; cannabis; epigenetics; granulosa cells; marijuana; Δ9-THC.
© The Author(s) 2022. Published by Oxford University Press on behalf of European Society of Human Reproduction and Embryology.
Figures

References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (November 2020, date last accessed).
-
- Balikova M, Hlozek T, Palenicek T, Tyls F, Viktorinova M, Melicher T, Androvicova R, Tomicek P, Roman M, Horacek J.. [Time profile of serum THC levels in occasional and chronic marihuana smokers after acute drug use—implication for driving motor vehicles]. Soud Lek 2014;59:2–6. - PubMed
-
- Battista N, Bari M, Maccarrone M.. Endocannabinoids and reproductive events in health and disease. Handb Exp Pharmacol 2015;231:341–365. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

