Concordance of SARS-CoV-2 RNA in Aerosols From a Nurses Station and in Nurses and Patients During a Hospital Ward Outbreak
- PMID: 35675074
- PMCID: PMC9178433
- DOI: 10.1001/jamanetworkopen.2022.16176
Concordance of SARS-CoV-2 RNA in Aerosols From a Nurses Station and in Nurses and Patients During a Hospital Ward Outbreak
Erratum in
-
Error in Data.JAMA Netw Open. 2022 Jun 1;5(6):e2221256. doi: 10.1001/jamanetworkopen.2022.21256. JAMA Netw Open. 2022. PMID: 35771583 Free PMC article. No abstract available.
Abstract
Importance: Aerosol-borne SARS-CoV-2 has not been linked specifically to nosocomial outbreaks.
Objective: To explore the genomic concordance of SARS-CoV-2 from aerosol particles of various sizes and infected nurses and patients during a nosocomial outbreak of COVID-19.
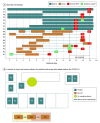
Design, setting, and participants: This cohort study included patients and nursing staff in a US Department of Veterans Affairs inpatient hospital unit and long-term-care facility during a COVID-19 outbreak between December 27, 2020, and January 8, 2021. Outbreak contact tracing was conducted using exposure histories and screening with reverse transcriptase-polymerase chain reaction (RT-PCR) for SARS-CoV-2. Size-selective particle samplers were deployed in diverse clinical areas of a multicampus health care system from November 2020 to March 2021. Viral genomic sequences from infected nurses and patients were sequenced and compared with ward nurses station aerosol samples.
Exposure: SARS-CoV-2.
Main outcomes and measures: The primary outcome was positive RT-PCR results and genomic similarity between SARS-CoV-2 RNA in aerosols and human samples. Air samplers were used to detect SARS-CoV-2 RNA in aerosols on hospital units where health care personnel were or were not under routine surveillance for SARS-CoV-2 infection.
Results: A total of 510 size-fractionated air particle samples were collected. Samples representing 3 size fractions (>10 μm, 2.5-10 μm, and <2.5 μm) obtained at the nurses station were positive for SARS-CoV-2 during the outbreak (3 of 30 samples [10%]) and negative during 9 other collection periods. SARS-CoV-2 partial genome sequences for the smallest particle fraction were 100% identical with all 3 human samples; the remaining size fractions shared >99.9% sequence identity with the human samples. Fragments of SARS-CoV-2 RNA were detected by RT-PCR in 24 of 300 samples (8.0%) in units where health care personnel were not under surveillance and 7 of 210 samples (3.3%; P = .03) where they were under surveillance.
Conclusions and relevance: In this cohort study, the finding of genetically identical SARS-CoV-2 RNA fragments in aerosols obtained from a nurses station and in human samples during a nosocomial outbreak suggests that aerosols may have contributed to hospital transmission. Surveillance, along with ventilation, masking, and distancing, may reduce the introduction of community-acquired SARS-CoV-2 into aerosols on hospital wards, thereby reducing the risk of hospital transmission.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

