Molecular Dynamics Simulations Establish the Molecular Basis for the Broad Allostery Hotspot Distributions in the Tetracycline Repressor
- PMID: 35675441
- PMCID: PMC9616627
- DOI: 10.1021/jacs.2c03275
Molecular Dynamics Simulations Establish the Molecular Basis for the Broad Allostery Hotspot Distributions in the Tetracycline Repressor
Abstract
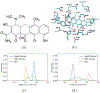
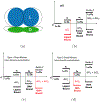
It is imperative to identify the network of residues essential to the allosteric coupling for the purpose of rationally engineering allostery in proteins. Deep mutational scanning analysis has emerged as a function-centric approach for identifying such allostery hotspots in a comprehensive and unbiased fashion, leading to observations that challenge our understanding of allostery at the molecular level. Specifically, a recent deep mutational scanning study of the tetracycline repressor (TetR) revealed an unexpectedly broad distribution of allostery hotspots throughout the protein structure. Using extensive molecular dynamics simulations (up to 50 μs) and free energy computations, we establish the molecular and energetic basis for the strong anticooperativity between the ligand and DNA binding sites. The computed free energy landscapes in different ligation states illustrate that allostery in TetR is well described by a conformational selection model, in which the apo state samples a broad set of conformations, and specific ones are selectively stabilized by either ligand or DNA binding. By examining a range of structural and dynamic properties of residues at both local and global scales, we observe that various analyses capture different subsets of experimentally identified hotspots, suggesting that these residues modulate allostery in distinct ways. These results motivate the development of a thermodynamic model that qualitatively explains the broad distribution of hotspot residues and their distinct features in molecular dynamics simulations. The multifaceted strategy that we establish here for hotspot evaluations and our insights into their mechanistic contributions are useful for modulating protein allostery in mechanistic and engineering studies.
Figures
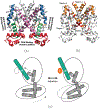

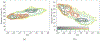
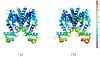
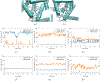


References
-
- Monod J; Wyman J; Changeux J-P On the nature of allosteric transitions: a plausible model. J. Mol. Biol 1965, 12, 88–118. - PubMed
-
- Koshland DEJ; Nemethy G; Filmer D Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochem. 1966, 5, 365–385. - PubMed
-
- Alberts B; Bray D; Lewis J; Raff M; Roberts K; Watson JD Molecular biology of the cell; Garland Publishing, Inc., 1994.
-
- Changeux J-P; Edelstein SJ Allosteric mechanisms of signal transduction. Science 2005, 308, 1424–1428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

