Comparison of CpG- and UpA-mediated restriction of RNA virus replication in mammalian and avian cells and investigation of potential ZAP-mediated shaping of host transcriptome compositions
- PMID: 35675984
- PMCID: PMC9297844
- DOI: 10.1261/rna.079102.122
Comparison of CpG- and UpA-mediated restriction of RNA virus replication in mammalian and avian cells and investigation of potential ZAP-mediated shaping of host transcriptome compositions
Abstract
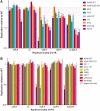
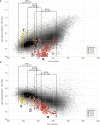
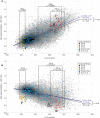
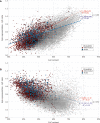
The ability of zinc finger antiviral protein (ZAP) to recognize and respond to RNA virus sequences with elevated frequencies of CpG dinucleotides has been proposed as a functional part of the vertebrate innate immune antiviral response. It has been further proposed that ZAP activity shapes compositions of cytoplasmic mRNA sequences to avoid self-recognition, particularly mRNAs for interferons (IFNs) and IFN-stimulated genes (ISGs) expressed during the antiviral state. We investigated whether restriction of the replication of mutants of influenza A virus (IAV) and the echovirus 7 (E7) replicon with high CpG and UpA frequencies varied in different species of mammals and birds. Cell lines from different bird orders showed substantial variability in restriction of CpG-high mutants of IAV and E7 replicons, whereas none restricted UpA-high mutants, in marked contrast to universal restriction of both mutants in mammalian cells. Dinucleotide representation in ISGs and IFN genes was compared with those of cellular transcriptomes to determine whether potential differences in inferred ZAP activity between species shaped dinucleotide compositions of highly expressed genes during the antiviral state. While mammalian type 1 IFN genes typically showed often profound suppression of CpG and UpA frequencies, there was no oversuppression of either in ISGs in any species, irrespective of their ability to restrict CpG- or UpA-high mutants. Similarly, genome sequences of mammalian and avian RNA viruses were compositionally equivalent, as were IAV strains recovered from ducks, chickens and humans. Overall, we found no evidence for host variability in inferred ZAP function shaping host or viral transcriptome compositions.
Keywords: RNA virus; dinucleotide; interferon; interferon-stimulated gene; trancriptome; zinc finger antiviral protein.
© 2022 Odon et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
- BB/N023803/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K004468/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011224/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- WT103767MA/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
