ATGL is a biosynthetic enzyme for fatty acid esters of hydroxy fatty acids
- PMID: 35676490
- PMCID: PMC9242854
- DOI: 10.1038/s41586-022-04787-x
ATGL is a biosynthetic enzyme for fatty acid esters of hydroxy fatty acids
Abstract
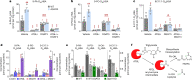
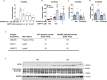
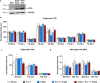
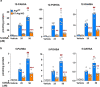
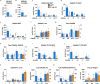
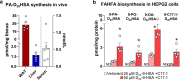
Branched fatty acid (FA) esters of hydroxy FAs (HFAs; FAHFAs) are recently discovered lipids that are conserved from yeast to mammals1,2. A subfamily, palmitic acid esters of hydroxy stearic acids (PAHSAs), are anti-inflammatory and anti-diabetic1,3. Humans and mice with insulin resistance have lower PAHSA levels in subcutaneous adipose tissue and serum1. PAHSA administration improves glucose tolerance and insulin sensitivity and reduces inflammation in obesity, diabetes and immune-mediated diseases1,4-7. The enzyme(s) responsible for FAHFA biosynthesis in vivo remains unknown. Here we identified adipose triglyceride lipase (ATGL, also known as patatin-like phospholipase domain containing 2 (PNPLA2)) as a candidate biosynthetic enzyme for FAHFAs using chemical biology and proteomics. We discovered that recombinant ATGL uses a transacylation reaction that esterifies an HFA with a FA from triglyceride (TG) or diglyceride to produce FAHFAs. Overexpression of wild-type, but not catalytically dead, ATGL increases FAHFA biosynthesis. Chemical inhibition of ATGL or genetic deletion of Atgl inhibits FAHFA biosynthesis and reduces the levels of FAHFA and FAHFA-TG. Levels of endogenous and nascent FAHFAs and FAHFA-TGs are 80-90 per cent lower in adipose tissue of mice in which Atgl is knocked out specifically in the adipose tissue. Increasing TG levels by upregulating diacylglycerol acyltransferase (DGAT) activity promotes FAHFA biosynthesis, and decreasing DGAT activity inhibits it, reinforcing TGs as FAHFA precursors. ATGL biosynthetic transacylase activity is present in human adipose tissue underscoring its potential clinical relevance. In summary, we discovered the first, to our knowledge, biosynthetic enzyme that catalyses the formation of the FAHFA ester bond in mammals. Whereas ATGL lipase activity is well known, our data establish a paradigm shift demonstrating that ATGL transacylase activity is biologically important.
© 2022. The Author(s).
Conflict of interest statement
B.B.K. and A. Saghatelian are inventors on patents WO2013166431A1 and WO2017070515A3, related to FA esters of HFAs and use of these lipids for the treatment of disease conditions. The submitted manuscript identifies an enzyme that can be targeted to increase the endogenous levels of these lipids.
Figures





References
MeSH terms
Substances
Grants and funding
- S10 OD021815/OD/NIH HHS/United States
- F30 DK112622/DK/NIDDK NIH HHS/United States
- R01 DK106210/DK/NIDDK NIH HHS/United States
- P30 DK046200/DK/NIDDK NIH HHS/United States
- K01 DK128075/DK/NIDDK NIH HHS/United States
- P30 DK057521/DK/NIDDK NIH HHS/United States
- R56 DK043051/DK/NIDDK NIH HHS/United States
- T32 DK007516/DK/NIDDK NIH HHS/United States
- P30 DK135043/DK/NIDDK NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
- T32 HL007374/HL/NHLBI NIH HHS/United States
- R01 DK043051/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

