Three-dimensional analysis of interstitial cells in the lamina propria of the murine vas deferens by confocal laser scanning microscopy and FIB/SEM
- PMID: 35676513
- PMCID: PMC9177838
- DOI: 10.1038/s41598-022-13245-7
Three-dimensional analysis of interstitial cells in the lamina propria of the murine vas deferens by confocal laser scanning microscopy and FIB/SEM
Abstract
The present study aimed to explore the three-dimensional (3D) ultrastructure of interstitial cells (ICs) within the lamina propria of the murine vas deferens and the spatial relationships between epithelial cells and surrounding cells. Focused ion beam scanning electron microscopy and confocal laser scanning microscopy were performed. ICs within the lamina propria had a flat, sheet-like structure of cytoplasm with multiple cellular processes. In addition, two types of 3D structures that comprised cell processes of flat, sheet-like ICs were observed: one was an accordion fold-like structure and the other was a rod-shaped structure. ICs were located parallel to the epithelium and were connected to each other via gap junctions or adherens junctions. Moreover, multiple sphere-shaped extracellular vesicle-like structures were frequently observed around the ICs. The ICs formed a complex 3D network comprising sheet-like cytoplasm and multiple cell processes with different 3D structures. From this morphological study, we noted that ICs within the lamina propria of murine vas deferens may be involved in signal transmission between the epithelium and smooth muscle cells by physical interaction and by exchanging extracellular vesicles.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

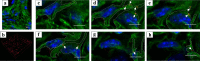
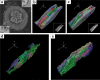


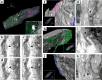
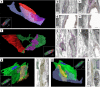
Similar articles
-
Three-Dimensional Analysis of Interstitial Cells in the Smooth Muscle Layer of Murine Vas Deferens Using Confocal Laser Scanning Microscopy and FIB/SEM.Microsc Microanal. 2022 Jan 26:1-9. doi: 10.1017/S1431927622000058. Online ahead of print. Microsc Microanal. 2022. PMID: 35078549
-
3D-electron microscopic characterization of interstitial cells in the human bladder upper lamina propria.Neurourol Urodyn. 2018 Jan;37(1):89-98. doi: 10.1002/nau.23270. Epub 2017 Mar 31. Neurourol Urodyn. 2018. PMID: 28370277
-
Identification of PDGFRα-positive interstitial cells in the distal segment of the murine vas deferens.Sci Rep. 2021 Apr 6;11(1):7553. doi: 10.1038/s41598-021-87049-6. Sci Rep. 2021. PMID: 33824385 Free PMC article.
-
Three-dimensional structure of the rat intestinal wall (mucosa and submucosa).Arch Histol Cytol. 1990 Mar;53(1):1-21. doi: 10.1679/aohc.53.1. Arch Histol Cytol. 1990. PMID: 2194550 Review.
-
Mucosa-Dependent, Stretch-Sensitive Spontaneous Activity in Seminal Vesicle.Adv Exp Med Biol. 2019;1124:217-231. doi: 10.1007/978-981-13-5895-1_9. Adv Exp Med Biol. 2019. PMID: 31183829 Review.
Cited by
-
Insights on Platelet-Derived Growth Factor Receptor α-Positive Interstitial Cells in the Male Reproductive Tract.Int J Mol Sci. 2024 Apr 8;25(7):4128. doi: 10.3390/ijms25074128. Int J Mol Sci. 2024. PMID: 38612936 Free PMC article. Review.
References
-
- Marberger H. The mechanisms of ejaculation. Basic Life Sci. 1974;4:99–110. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

