This is a preprint.
Imprinted antibody responses against SARS-CoV-2 Omicron sublineages
- PMID: 35677069
- PMCID: PMC9176643
- DOI: 10.1101/2022.05.08.491108
Imprinted antibody responses against SARS-CoV-2 Omicron sublineages
Update in
-
Imprinted antibody responses against SARS-CoV-2 Omicron sublineages.Science. 2022 Nov 11;378(6620):619-627. doi: 10.1126/science.adc9127. Epub 2022 Oct 20. Science. 2022. PMID: 36264829
Abstract
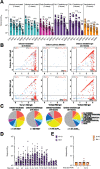
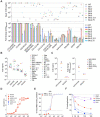
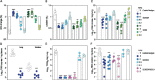
SARS-CoV-2 Omicron sublineages carry distinct spike mutations and represent an antigenic shift resulting in escape from antibodies induced by previous infection or vaccination. We show that hybrid immunity or vaccine boosters result in potent plasma neutralizing activity against Omicron BA.1 and BA.2 and that breakthrough infections, but not vaccination-only, induce neutralizing activity in the nasal mucosa. Consistent with immunological imprinting, most antibodies derived from memory B cells or plasma cells of Omicron breakthrough cases cross-react with the Wuhan-Hu-1, BA.1 and BA.2 receptor-binding domains whereas Omicron primary infections elicit B cells of narrow specificity. While most clinical antibodies have reduced neutralization of Omicron, we identified an ultrapotent pan-variant antibody, that is unaffected by any Omicron lineage spike mutations and is a strong candidate for clinical development.
Conflict of interest statement
Competing interests: D.P., Ad.M., F.Z., M.G., C.S.F., J.B., C.S., H.V.D., K.H., W.R., M.A.S., G.Sc., B.G., F.B., J.d.I., A.R., J.Z., N.F., H.K., M.M.R, J.N., F.A.L., G.S., L.P., H.W.V., A.L., M.S.P. and D.C. are employees of Vir Biotechnology Inc. and may hold shares in Vir Biotechnology Inc. L.A.P. is a former employee and shareholder in Regeneron Pharmaceuticals. Regeneron provided no funding for this work. H.W.V. is a founder and holds shares in PierianDx and Casma Therapeutics. Neither company provided resources. D.C. is currently listed as an inventor on multiple patent applications, which disclose the subject matter described in this manuscript. The Veesler laboratory has received a sponsored research agreement from Vir Biotechnology Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Viana R., Moyo S., Amoako D. G., Tegally H., Scheepers C., Althaus C. L., Anyaneji U. J., Bester P. A., Boni M. F., Chand M., Choga W. T., Colquhoun R., Davids M., Deforche K., Doolabh D., du Plessis L., Engelbrecht S., Everatt J., Giandhari J., Giovanetti M., Hardie D., Hill V., Hsiao N.-Y., Iranzadeh A., Ismail A., Joseph C., Joseph R., Koopile L., Kosakovsky Pond S. L., Kraemer M. U. G., Kuate-Lere L., Laguda-Akingba O., Lesetedi-Mafoko O., Lessells R. J., Lockman S., Lucaci A. G., Maharaj A., Mahlangu B., Maponga T., Mahlakwane K., Makatini Z., Marais G., Maruapula D., Masupu K., Matshaba M., Mayaphi S., Mbhele N., Mbulawa M. B., Mendes A., Mlisana K., Mnguni A., Mohale T., Moir M., Moruisi K., Mosepele M., Motsatsi G., Motswaledi M. S., Mphoyakgosi T., Msomi N., Mwangi P. N., Naidoo Y., Ntuli N., Nyaga M., Olubayo L., Pillay S., Radibe B., Ramphal Y., Ramphal U., San J. E., Scott L., Shapiro R., Singh L., Smith-Lawrence P., Stevens W., Strydom A., Subramoney K., Tebeila N., Tshiabuila D., Tsui J., van Wyk S., Weaver S., Wibmer C. K., Wilkinson E., Wolter N., Zarebski A. E., Zuze B., Goedhals D., Preiser W., Treurnicht F., Venter M., Williamson C., Pybus O. G., Bhiman J., Glass A., Martin D. P., Rambaut A., Gaseitsiwe S., von Gottberg A., de Oliveira T., Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature (2022), doi: 10.1038/d41586-021-03832-5. - DOI - PMC - PubMed
-
- Yu J., Collier A.-R. Y., Rowe M., Mardas F., Ventura J. D., Wan H., Miller J., Powers O., Chung B., Siamatu M., Hachmann N. P., Surve N., Nampanya F., Chandrashekar A., Barouch D. H., Comparable neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 variants. medRxiv (2022), doi: 10.1101/2022.02.06.22270533. - DOI - PMC - PubMed
-
- Cameroni E., Bowen J. E., Rosen L. E., Saliba C., Zepeda S. K., Culap K., Pinto D., VanBlargan L. A., De Marco A., di Iulio J., Zatta F., Kaiser H., Noack J., Farhat N., Czudnochowski N., Havenar-Daughton C., Sprouse K. R., Dillen J. R., Powell A. E., Chen A., Maher C., Yin L., Sun D., Soriaga L., Bassi J., Silacci-Fregni C., Gustafsson C., Franko N. M., Logue J., Iqbal N. T., Mazzitelli I., Geffner J., Grifantini R., Chu H., Gori A., Riva A., Giannini O., Ceschi A., Ferrari P., Cippà P. E., Franzetti-Pellanda A., Garzoni C., Halfmann P. J., Kawaoka Y., Hebner C., Purcell L. A., Piccoli L., Pizzuto M. S., Walls A. C., Diamond M. S., Telenti A., Virgin H. W., Lanzavecchia A., Snell G., Veesler D., Corti D., Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature (2021), doi: 10.1038/d41586-021-03825-4. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
