Classification of Muscle Invasive Bladder Cancer to Predict Prognosis of Patients Treated with Immunotherapy
- PMID: 35677536
- PMCID: PMC9170513
- DOI: 10.1155/2022/6737241
Classification of Muscle Invasive Bladder Cancer to Predict Prognosis of Patients Treated with Immunotherapy
Abstract
Background: Recently, immunotherapies have been approved for advanced muscle invasive bladder cancer (MIBC) treatment, but only a small fraction of MIBC patients could achieve a durable drug response. Our study is aimed at identifying tumor microenvironment (TME) subtypes that have different immunotherapy response rates.
Methods: The mRNA expression profiles of MIBC samples from seven discovery datasets (GSE13507, GSE31684, GSE32548, GSE32894, GSE48075, GSE48276, and GSE69795) were analyzed to identify TME subtypes. The identified TME subtypes were then validated by an independent dataset (TCGA-MIBC). The subtype-related biomarkers were discovered using computational analyses and then utilized to establish a random forest predictive model. The associations of TME subtypes with immunotherapy therapeutic responses were investigated in a group of patients who had been treated with immunotherapy. A prognostic index model was constructed using the subtype-related biomarkers. Two nomograms were built by the subtype-related biomarkers or the clinical parameters.
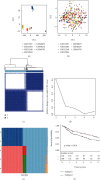
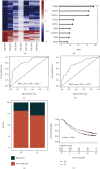
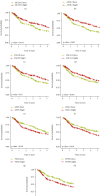
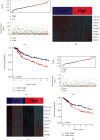
Results: Two TME subtypes, including ECM-enriched class (EC) and immune-enriched class (IC), were found. EC was associated with greater extracellular matrix (ECM) pathways, and IC was correlated with immune pathways, respectively. Overall survival was significantly greater for tumors classified as IC, whereas the EC subtype had a worse prognosis. A total of nine genes (AKAP12, APOL3, CXCL13, CXCL9, GBP4, LRIG1, PEG3, PODN, and PTPRD) were selected by computational analyses to construct the random forest model. The area under the curve (AUC) values for this model were 0.827 and 0.767 in the testing and external validation datasets, respectively. Therapeutic response rates were greater in IC patients than in EC patients (28 percent vs. 18 percent). Patients with a high prognostic index had a poorer prognosis than those with a low prognostic index. The nomogram constructed from nine genes and stage achieved a C-index of 0.71.
Conclusion: The present investigation defined two distinct TME subtypes and developed models to assess immunotherapeutic treatment outcomes.
Copyright © 2022 Zhifeng Wang et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

