Genetic background and sex control the outcome of high-fat diet feeding in mice
- PMID: 35677645
- PMCID: PMC9167980
- DOI: 10.1016/j.isci.2022.104468
Genetic background and sex control the outcome of high-fat diet feeding in mice
Abstract
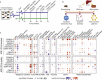
The sharp increase in obesity prevalence worldwide is mainly attributable to changes in physical activity and eating behavior but the metabolic and clinical impacts of these obesogenic conditions vary between sexes and genetic backgrounds. This warrants personalized treatments of obesity and its complications, which require a thorough understanding of the diversity of metabolic responses to high-fat diet intake. By analyzing nine genetically diverse mouse strains, we show that much like humans, mice exhibit a huge variety of physiological and biochemical responses to high-fat diet. The strains exhibit various degrees of alterations in their phenotypic makeup. At the transcriptome level, we observe dysregulations of immunity, translation machinery, and mitochondrial genes. At the biochemical level, the enzymatic activity of mitochondrial complexes is affected. The diversity across mouse strains, diets, and sexes parallels that found in humans and supports the use of diverse mouse populations in future mechanistic or preclinical studies on metabolic dysfunctions.
Keywords: Biological sciences; Endocrinology; Obesity medicine.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Alhindi Y., Vaanholt L.M., Al-Tarrah M., Gray S.R., Speakman J.R., Hambly C., Alanazi B.S., Gabriel B.M., Lionikas A., Ratkevicius A. Low citrate synthase activity is associated with glucose intolerance and lipotoxicity. J. Nutr. Metab. 2019;2019:1–14. doi: 10.1155/2019/8594825. - DOI - PMC - PubMed
-
- Atsumi A., Ueda K., Irie F., Sairenchi T., Iimura K., Watanabe H., Iso H., Ota H., Aonuma K. Relationship between cold temperature and cardiovascular mortality, with assessment of effect modification by individual characteristics. Circ. J. 2013;77:1854–1861. doi: 10.1253/circj.cj-12-0916. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

