Deep Learning in RNA Structure Studies
- PMID: 35677883
- PMCID: PMC9168262
- DOI: 10.3389/fmolb.2022.869601
Deep Learning in RNA Structure Studies
Abstract
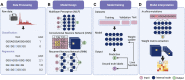
Deep learning, or artificial neural networks, is a type of machine learning algorithm that can decipher underlying relationships from large volumes of data and has been successfully applied to solve structural biology questions, such as RNA structure. RNA can fold into complex RNA structures by forming hydrogen bonds, thereby playing an essential role in biological processes. While experimental effort has enabled resolving RNA structure at the genome-wide scale, deep learning has been more recently introduced for studying RNA structure and its functionality. Here, we discuss successful applications of deep learning to solve RNA problems, including predictions of RNA structures, non-canonical G-quadruplex, RNA-protein interactions and RNA switches. Following these cases, we give a general guide to deep learning for solving RNA structure problems.
Keywords: RNA G-quadruplex; RNA secondary structure; RNA structure prediction; RNA tertiary structure; RNA-protein interaction; deep learning.
Copyright © 2022 Yu, Qi and Ding.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

