Normalization techniques for high-throughput screening by infrared matrix-assisted laser desorption electrospray ionization mass spectrometry
- PMID: 35678360
- PMCID: PMC9287052
- DOI: 10.1002/jms.4869
Normalization techniques for high-throughput screening by infrared matrix-assisted laser desorption electrospray ionization mass spectrometry
Abstract
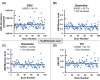
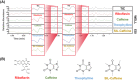
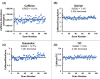
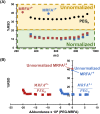
Mass spectrometry (MS) is an effective analytical tool for high-throughput screening (HTS) in the drug discovery field. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) MS is a high-throughput platform that has achieved analysis times of sub-seconds-per-sample. Due to the high-throughput analysis speed, methods are needed to increase the analyte signal while decreasing the variability in IR-MALDESI-MS analyses to improve data quality and reduce false-positive hits. The Z-factor is used as a statistic of assay quality that can be improved by reducing the variation of target ion abundances or increasing signal. Herein we report optimal solvent compositions for increasing measured analyte abundances with direct analysis by IR-MALDESI-MS. We also evaluate normalization strategies, such as adding a normalization standard that is similar or dissimilar in structure to the model target drug, to reduce the variability of measured analyte abundances with direct analyses by IR-MALDESI-MS in both positive and negative ionization modes.
Keywords: IR-MALDESI; Orbitrap mass spectrometer; high-throughput screening; normalization.
© 2022 The Authors. Journal of Mass Spectrometry published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Next-Generation Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Source for Mass Spectrometry Imaging and High-Throughput Screening.J Am Soc Mass Spectrom. 2022 Nov 2;33(11):2070-2077. doi: 10.1021/jasms.2c00178. Epub 2022 Sep 29. J Am Soc Mass Spectrom. 2022. PMID: 36173393 Free PMC article.
-
Optimized C-Trap Timing of an Orbitrap 240 Mass Spectrometer for High-Throughput Screening and Native MS by IR-MALDESI.J Am Soc Mass Spectrom. 2022 Feb 2;33(2):328-334. doi: 10.1021/jasms.1c00319. Epub 2022 Jan 24. J Am Soc Mass Spectrom. 2022. PMID: 35073091 Free PMC article.
-
Ultra-High-Throughput Ambient MS: Direct Analysis at 22 Samples per Second by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry.Anal Chem. 2022 Mar 29;94(12):4913-4918. doi: 10.1021/acs.analchem.1c04605. Epub 2022 Mar 15. Anal Chem. 2022. PMID: 35290016
-
The development and application of matrix assisted laser desorption electrospray ionization: The teenage years.Mass Spectrom Rev. 2023 Jan;42(1):35-66. doi: 10.1002/mas.21696. Epub 2021 May 24. Mass Spectrom Rev. 2023. PMID: 34028071 Free PMC article. Review.
-
Surface analysis of lipids by mass spectrometry: more than just imaging.Prog Lipid Res. 2013 Oct;52(4):329-53. doi: 10.1016/j.plipres.2013.04.005. Epub 2013 Apr 24. Prog Lipid Res. 2013. PMID: 23623802 Review.
Cited by
-
DISC-3D: dual-hydrogel system enhances optical imaging and enables correlative mass spectrometry imaging of invading multicellular tumor spheroids.Sci Rep. 2023 Jul 31;13(1):12383. doi: 10.1038/s41598-023-38699-1. Sci Rep. 2023. PMID: 37524722 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

