The CRISPR-Cas System Differentially Regulates Surface-Attached and Pellicle Biofilm in Salmonella enterica Serovar Typhimurium
- PMID: 35678575
- PMCID: PMC9241790
- DOI: 10.1128/spectrum.00202-22
The CRISPR-Cas System Differentially Regulates Surface-Attached and Pellicle Biofilm in Salmonella enterica Serovar Typhimurium
Abstract
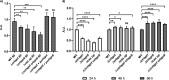
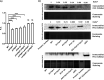
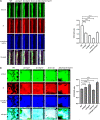
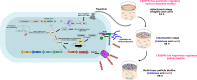
The CRISPR-Cas mediated regulation of biofilm by Salmonella enterica serovar Typhimurium was investigated by deleting CRISPR-Cas components ΔcrisprI, ΔcrisprII, ΔΔcrisprI crisprII, and Δcas op. We determined that the system positively regulates surface biofilm while inhibiting pellicle biofilm formation. Results of real-time PCR suggest that the flagellar (fliC, flgK) and curli (csgA) genes were repressed in knockout strains, causing reduced surface biofilm. The mutants displayed altered pellicle biofilm architecture. They exhibited bacterial multilayers and a denser extracellular matrix with enhanced cellulose and less curli, ergo weaker pellicles than those of the wild type. The cellulose secretion was more in the knockout strains due to the upregulation of bcsC, which is necessary for cellulose export. We hypothesized that the secreted cellulose quickly integrates into the pellicle, leading to enhanced pellicular cellulose in the knockout strains. We determined that crp is upregulated in the knockout strains, thereby inhibiting the expression of csgD and, hence, also of csgA and bcsA. The conflicting upregulation of bcsC, the last gene of the bcsABZC operon, could be caused by independent regulation by the CRISPR-Cas system owing to a partial match between the CRISPR spacers and bcsC gene. The cAMP-regulated protein (CRP)-mediated regulation of the flagellar genes in the knockout strains was probably circumvented through the regulation of yddx governing the availability of the sigma factor σ28 that further regulates class 3 flagellar genes (fliC, fljB, and flgK). Additionally, the variations in the lipopolysaccharide (LPS) profile and expression of LPS-related genes (rfaC, rfbG, and rfbI) in knockout strains could also contribute to the altered pellicle architecture. Collectively, we establish that the CRISPR-Cas system differentially regulates the formation of surface-attached and pellicle biofilm. IMPORTANCE In addition to being implicated in bacterial immunity and genome editing, the CRISPR-Cas system has recently been demonstrated to regulate endogenous gene expression and biofilm formation. While the function of individual cas genes in controlling Salmonella biofilm has been explored, the regulatory role of CRISPR arrays in biofilm is less studied. Moreover, studies have focused on the effects of the CRISPR-Cas system on surface-associated biofilms, and comprehensive studies on the impact of the system on pellicle biofilm remain an unexplored niche. We demonstrate that the CRISPR array and cas genes modulate the expression of various biofilm genes in Salmonella, whereby surface and pellicle biofilm formation is distinctively regulated.
Keywords: Salmonella; pellicle biofilm; surface-attached biofilm; type I-E CRISPR-Cas system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

