Protocol for Increasing the Sensitivity of MS-Based Protein Detection in Human Chorionic Villi
- PMID: 35678669
- PMCID: PMC9164042
- DOI: 10.3390/cimb44050140
Protocol for Increasing the Sensitivity of MS-Based Protein Detection in Human Chorionic Villi
Abstract
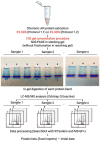
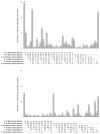
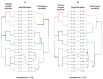
An important step in the proteomic analysis of missing proteins is the use of a wide range of tissues, optimal extraction, and the processing of protein material in order to ensure the highest sensitivity in downstream protein detection. This work describes a purification protocol for identifying low-abundance proteins in human chorionic villi using the proposed "1DE-gel concentration" method. This involves the removal of SDS in a short electrophoresis run in a stacking gel without protein separation. Following the in-gel digestion of the obtained holistic single protein band, we used the peptide mixture for further LC-MS/MS analysis. Statistically significant results were derived from six datasets, containing three treatments, each from two tissue sources (elective or missed abortions). The 1DE-gel concentration increased the coverage of the chorionic villus proteome. Our approach allowed the identification of 15 low-abundance proteins, of which some had not been previously detected via the mass spectrometry of trophoblasts. In the post hoc data analysis, we found a dubious or uncertain protein (PSG7) encoded on human chromosome 19 according to neXtProt. A proteomic sample preparation workflow with the 1DE-gel concentration can be used as a prospective tool for uncovering the low-abundance part of the human proteome.
Keywords: 1DE-gel concentration; LC–MS/MS; PSG7; SDS extracts; bioinformatics; chorionic villi; elective abortion; low-abundance proteins; missed abortion.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

