Alchemical Free Energy Calculations to Investigate Protein-Protein Interactions: the Case of the CDC42/PAK1 Complex
- PMID: 35679463
- PMCID: PMC9241073
- DOI: 10.1021/acs.jcim.2c00348
Alchemical Free Energy Calculations to Investigate Protein-Protein Interactions: the Case of the CDC42/PAK1 Complex
Abstract
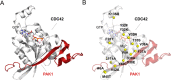
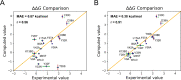
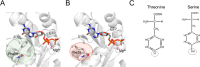
Here, we show that alchemical free energy calculations can quantitatively compute the effect of mutations at the protein-protein interface. As a test case, we have used the protein complex formed by the small Rho-GTPase CDC42 and its downstream effector PAK1, a serine/threonine kinase. Notably, the CDC42/PAK1 complex offers a wealth of structural, mutagenesis, and binding affinity data because of its central role in cellular signaling and cancer progression. In this context, we have considered 16 mutations in the CDC42/PAK1 complex and obtained excellent agreement between computed and experimental data on binding affinity. Importantly, we also show that a careful analysis of the side-chain conformations in the mutated amino acids can considerably improve the computed estimates, solving issues related to sampling limitations. Overall, this study demonstrates that alchemical free energy calculations can conveniently be integrated into the design of experimental mutagenesis studies.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Borgia A.; Borgia M. B.; Bugge K.; Kissling V. M.; Heidarsson P. O.; Fernandes C. B.; Sottini A.; Soranno A.; Buholzer K. J.; Nettels D.; Kragelund B. B.; Best R. B.; Schuler B. Extreme Disorder in an Ultrahigh-Affinity Protein Complex. Nature 2018, 555, 61–66. 10.1038/nature25762. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

