Heterogeneous data integration methods for patient similarity networks
- PMID: 35679533
- PMCID: PMC9294435
- DOI: 10.1093/bib/bbac207
Heterogeneous data integration methods for patient similarity networks
Abstract
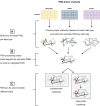
Patient similarity networks (PSNs), where patients are represented as nodes and their similarities as weighted edges, are being increasingly used in clinical research. These networks provide an insightful summary of the relationships among patients and can be exploited by inductive or transductive learning algorithms for the prediction of patient outcome, phenotype and disease risk. PSNs can also be easily visualized, thus offering a natural way to inspect complex heterogeneous patient data and providing some level of explainability of the predictions obtained by machine learning algorithms. The advent of high-throughput technologies, enabling us to acquire high-dimensional views of the same patients (e.g. omics data, laboratory data, imaging data), calls for the development of data fusion techniques for PSNs in order to leverage this rich heterogeneous information. In this article, we review existing methods for integrating multiple biomedical data views to construct PSNs, together with the different patient similarity measures that have been proposed. We also review methods that have appeared in the machine learning literature but have not yet been applied to PSNs, thus providing a resource to navigate the vast machine learning literature existing on this topic. In particular, we focus on methods that could be used to integrate very heterogeneous datasets, including multi-omics data as well as data derived from clinical information and medical imaging.
Keywords: biomedical applications; data fusion; multimodal data; patient similarity networks.
© The Author(s) 2022. Published by Oxford University Press.
Figures



References
Publication types
MeSH terms
Grants and funding
- BB/F00964X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M025047/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K004131/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/T001070/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

