RNase H-based analysis of synthetic mRNA 5' cap incorporation
- PMID: 35680168
- PMCID: PMC9297845
- DOI: 10.1261/rna.079173.122
RNase H-based analysis of synthetic mRNA 5' cap incorporation
Abstract
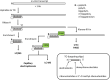
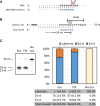
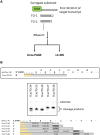
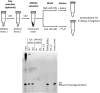
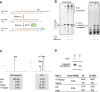
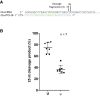
Advances in mRNA synthesis and lipid nanoparticles technologies have helped make mRNA therapeutics and vaccines a reality. The 5' cap structure is a crucial modification required to functionalize synthetic mRNA for efficient protein translation in vivo and evasion of cellular innate immune responses. The extent of 5' cap incorporation is one of the critical quality attributes in mRNA manufacturing. RNA cap analysis involves multiple steps: generation of predefined short fragments from the 5' end of the kilobase-long synthetic mRNA molecules using RNase H, a ribozyme or a DNAzyme, enrichment of the 5' cleavage products, and LC-MS intact mass analysis. In this paper, we describe (1) a framework to design site-specific RNA cleavage using RNase H; (2) a method to fluorescently label the RNase H cleavage fragments for more accessible readout methods such as gel electrophoresis or high-throughput capillary electrophoresis; (3) a simplified method for post-RNase H purification using desthiobiotinylated oligonucleotides and streptavidin magnetic beads followed by elution using water. By providing a design framework for RNase H-based RNA 5' cap analysis using less resource-intensive analytical methods, we hope to make RNA cap analysis more accessible to the scientific community.
Keywords: 5′ cap; LC-MS; RNA cap analysis; RNA capping; RNase H; synthetic mRNA.
© 2022 Chan et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
