Interferon-α-mediated therapeutic resistance in early rheumatoid arthritis implicates epigenetic reprogramming
- PMID: 35680389
- PMCID: PMC9380486
- DOI: 10.1136/annrheumdis-2022-222370
Interferon-α-mediated therapeutic resistance in early rheumatoid arthritis implicates epigenetic reprogramming
Abstract
Objectives: An interferon (IFN) gene signature (IGS) is present in approximately 50% of early, treatment naive rheumatoid arthritis (eRA) patients where it has been shown to negatively impact initial response to treatment. We wished to validate this effect and explore potential mechanisms of action.
Methods: In a multicentre inception cohort of eRA patients (n=191), we examined the whole blood IGS (MxA, IFI44L, OAS1, IFI6, ISG15) with reference to circulating IFN proteins, clinical outcomes and epigenetic influences on circulating CD19+ B and CD4+ T lymphocytes.
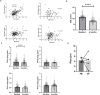
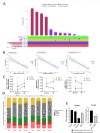
Results: We reproduced our previous findings demonstrating a raised baseline IGS. We additionally showed, for the first time, that the IGS in eRA reflects circulating IFN-α protein. Paired longitudinal analysis demonstrated a significant reduction between baseline and 6-month IGS and IFN-α levels (p<0.0001 for both). Despite this fall, a raised baseline IGS predicted worse 6-month clinical outcomes such as increased disease activity score (DAS-28, p=0.025) and lower likelihood of a good EULAR clinical response (p=0.034), which was independent of other conventional predictors of disease activity and clinical response. Molecular analysis of CD4+ T cells and CD19+ B cells demonstrated differentially methylated CPG sites and dysregulated expression of disease relevant genes, including PARP9, STAT1, and EPSTI1, associated with baseline IGS/IFNα levels. Differentially methylated CPG sites implicated altered transcription factor binding in B cells (GATA3, ETSI, NFATC2, EZH2) and T cells (p300, HIF1α).
Conclusions: Our data suggest that, in eRA, IFN-α can cause a sustained, epigenetically mediated, pathogenic increase in lymphocyte activation and proliferation, and that the IGS is, therefore, a robust prognostic biomarker. Its persistent harmful effects provide a rationale for the initial therapeutic targeting of IFN-α in selected patients with eRA.
Keywords: antirheumatic agents; arthritis, rheumatoid; immune system diseases; inflammation.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: JDI discloses research grants from Pfizer, Janssen and GSK; conference support from Eli Lilly and Gilead; speaker/consulting fees from AbbVie, BMS, Gilead, Roche and UCB. FAHC discloses speaker fees from AstraZeneca.The remaining authors have no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

