Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment
- PMID: 35680610
- PMCID: PMC9347785
- DOI: 10.1002/jmv.27927
Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment
Abstract
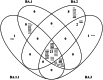
The Omicron variant of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has now spread throughout the world. We used computational tools to assess the spike infectivity, transmission, and pathogenicity of Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) in this study. BA.1 has 39 mutations, BA.1.1 has 40 mutations, BA.2 has 31 mutations, and BA.3 has 34 mutations, with 21 shared mutations between all. We observed 11 common mutations in Omicron's receptor-binding domain (RBD) and sub-variants. In pathogenicity analysis, the Y505H, N786K, T95I, N211I, N856K, and V213R mutations in omicron and sub-variants are predicted to be deleterious. Due to the major effect of the mutations characterizing in the RBD, we found that Omicron and sub-variants had a higher positive electrostatic surface potential. This could increase interaction between RBD and negative electrostatic surface potential human angiotensin-converting enzyme 2 (hACE2). Omicron and sub-variants had a higher affinity for hACE2 and the potential for increased transmission when compared to the wild-type (WT). Negative electrostatic potential of N-terminal domain (NTD) of the spike protein value indicates that the Omicron variant binds receptors less efficiently than the WT. Given that at least one receptor is highly expressed in lung and bronchial cells, the electrostatic potential of NTD negative value could be one of the factors contributing to why the Omicron variant is thought to be less harmful to the lower respiratory tract. Among Omicron sub-lineages, BA.2 and BA.3 have a higher transmission potential than BA.1 and BA.1.1. We predicted that mutated residues in BA.1.1 (K478), BA.2 (R400, R490, and R495), and BA.3 (R397 and H499) formation of new salt bridges and hydrogen bonds. Omicron and sub-variant mutations at Receptor-binding Motif (RBM) residues such as Q493R, N501Y, Q498, T478K, and Y505H all contribute significantly to binding affinity with human ACE2. Interactions with Omicron variant mutations at residues 493, 496, 498, and 501 seem to restore ACE2 binding effectiveness lost due to other mutations like K417N.
Keywords: BA.1; BA.2; BA.3; BA1.1; COVID-19; Omicron; SARS-CoV-2.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Wang L, Cheng G. Sequence analysis of the emerging SARS‐CoV‐2 variant Omicron in South Africa. J Med Virol. 2022;94(4):1728‐1733 - PubMed
-
- Kumar S. COVID‐19: a drug repurposing and biomarker identification by using comprehensive gene‐disease associations through protein‐protein interaction network analysis . Preprints. 2020;2020030440.
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

