Mitigation of SARS-CoV-2 transmission at a large public university
- PMID: 35680861
- PMCID: PMC9184485
- DOI: 10.1038/s41467-022-30833-3
Mitigation of SARS-CoV-2 transmission at a large public university
Abstract
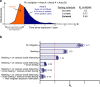
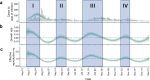
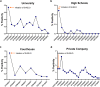
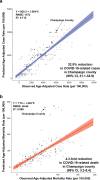
In Fall 2020, universities saw extensive transmission of SARS-CoV-2 among their populations, threatening health of the university and surrounding communities, and viability of in-person instruction. Here we report a case study at the University of Illinois at Urbana-Champaign, where a multimodal "SHIELD: Target, Test, and Tell" program, with other non-pharmaceutical interventions, was employed to keep classrooms and laboratories open. The program included epidemiological modeling and surveillance, fast/frequent testing using a novel low-cost and scalable saliva-based RT-qPCR assay for SARS-CoV-2 that bypasses RNA extraction, called covidSHIELD, and digital tools for communication and compliance. In Fall 2020, we performed >1,000,000 covidSHIELD tests, positivity rates remained low, we had zero COVID-19-related hospitalizations or deaths amongst our university community, and mortality in the surrounding Champaign County was reduced more than 4-fold relative to expected. This case study shows that fast/frequent testing and other interventions mitigated transmission of SARS-CoV-2 at a large public university.
© 2022. The Author(s).
Conflict of interest statement
University of Illinois at Urbana-Champaign has filed two pending patent applications related to the saliva-based assay described herein (US Nonprovisional Application No. 17/350,670, filed June 17, 2021; US National Phase Application No. 17/636,469, filed on February 18, 2022), and D.R.E.R., R.L.H., F.G.A., K.J.G., L.W., C.R.B., M.D.B., T.M.F., and P.J.H. are co-inventors. S.J.P., J.M.P., W.C.S., R.J.J., and A.C.C. are associated with Rokwire, which created the Safer Illinois App. G.D. and N.W. are associated with TekMill which created some of the automation used in the VDL COVID-19 Testing Lab at UIUC. P.J.H., A.C.C., S.A.M. and J.T.W. are Members of the Board of Managers for the University Related Organization SHIELD T3, LLC, which has obtained a non-exclusive license to this technology. The remaining authors declare no competing interests.
Figures



References
-
- Richmond, C. S., Sabin, A. P., Jobe, D. A., Lovrich, S. D. & Kenny, P. A. SARS-CoV-2 sequencing reveals rapid transmission from college student clusters resulting in morbidity and deaths in vulnerable populations. Preprint at medRxiv10.1101/2020.10.12.20210294 (2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

